ASRB NET 2025 Paper Discussion | Full Solution + Analysis | Agriculture / Plant Breeding / Genetics / Seed Science
Welcome to the most accurate and detailed ASRB NET 2025 Paper Discussion, presented by Satyam Sir, based on candidate recall, expert evaluation, and authentic question pattern analysis.
📌 What’s Covered in This Video?
✔ Complete paper-wise discussion (Part A + Part B + Part C)
✔ Correct answer key for all sections
✔ Difficulty level analysis
✔ Chapter-wise weightage
✔ High-scoring topics of 2025
✔ Expected cutoff prediction (General / OBC / SC / ST / EWS)
✔ Section-wise MCQ breakup
✔ Strategy to prepare for NET 2026🎯 Maha Marathon - 6 | Plant Breeding | Unit-2: Domestication & Germplasm Conservation | By Satyam Sharma 🎓
Welcome to Maha Marathon - 6 of the Plant Breeding Series by Satyam Sharma (Ph.D. Scholar, IARI New Delhi) —
ICAR JRF / SRF | ARS | MSc / PhD Entrances | State Exams | Agriculture Aspirants
#PlantBreeding #Domestication #GermplasmConservation #SatyamSharma #AgricultureExams #IARI #ICARJRF #ICARSRF #Genetics #MahaMarathon #PlantGeneticResourcesAfter months of teaching with full dedication, I’ve decided to start a new and transparent journey in Plant Science education.
Explained by: Satyam Sharma (AIR-5, JRF Plant Science)
Course: Plant Sciences Master Course – Target Batch 2026
Subscribe for more conceptual videos on Plant Breeding, Genetics, Biotechnology & Seed Science
ASRB NET 2025 Question Paper Analysis
• Moderate difficulty
• Maximum weightage from:
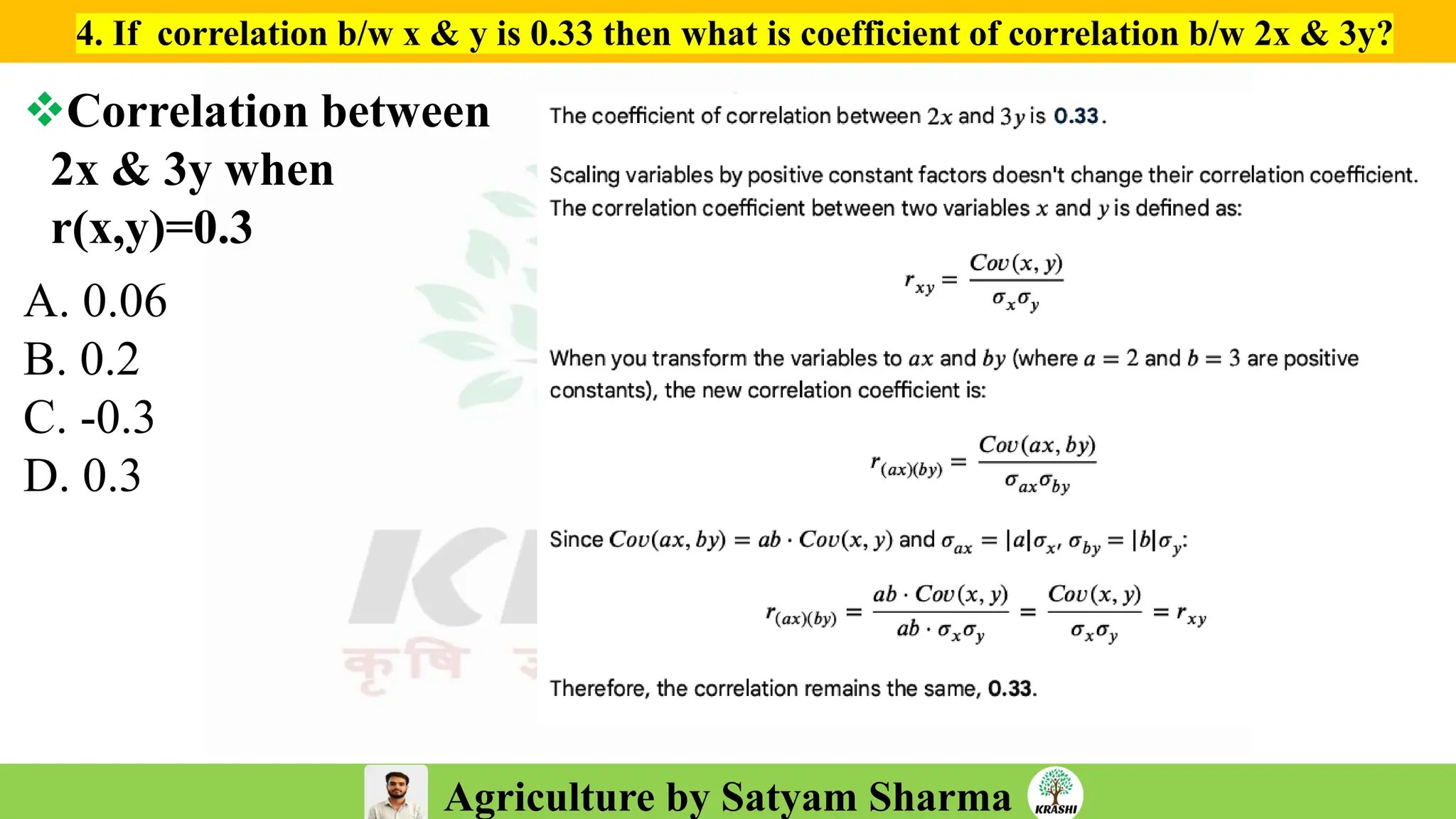
– Numerical aptitude
– Logical reasoning
– Current agriculture
• Mostly moderate-to-tough
• High weightage from:
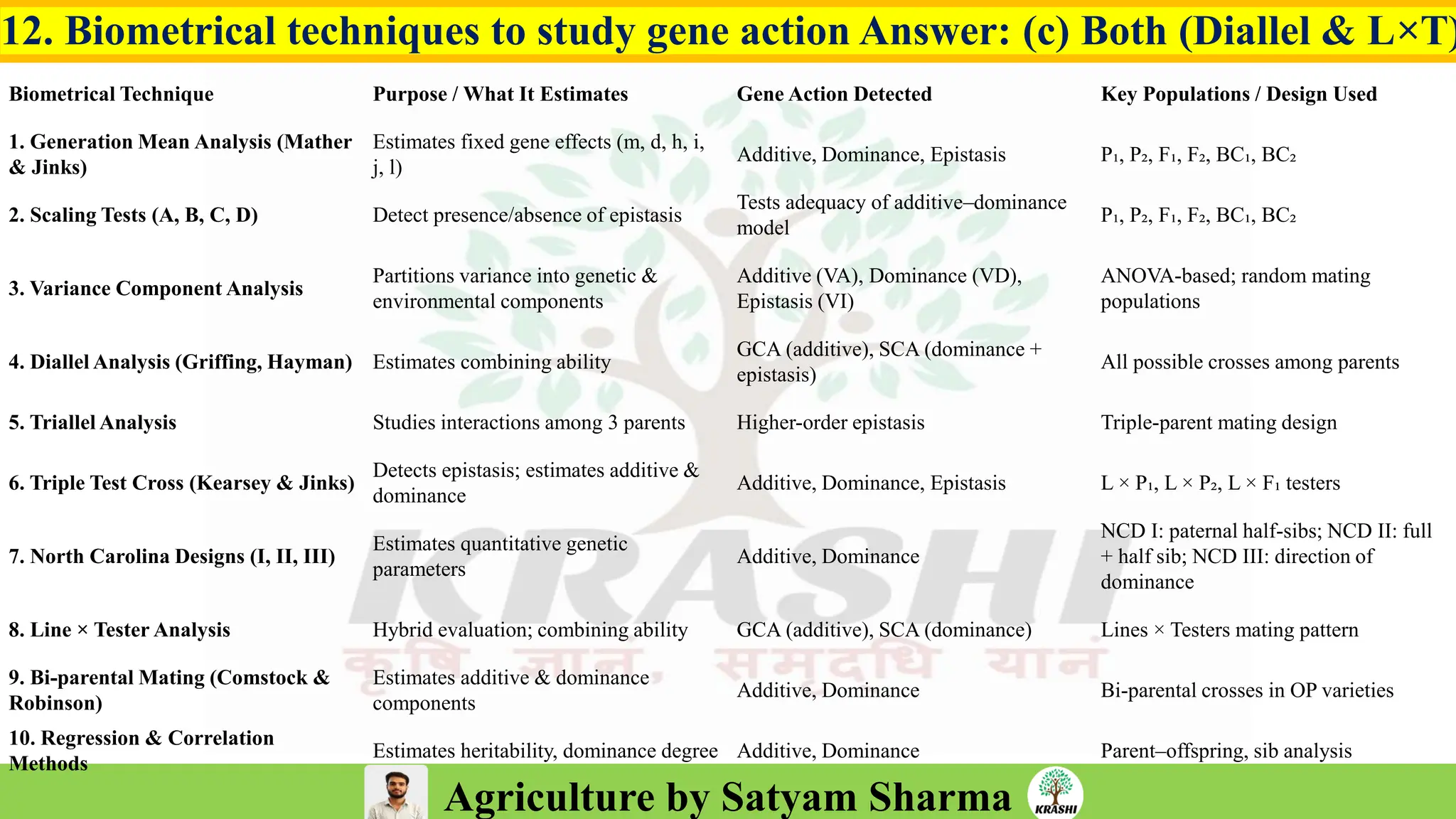
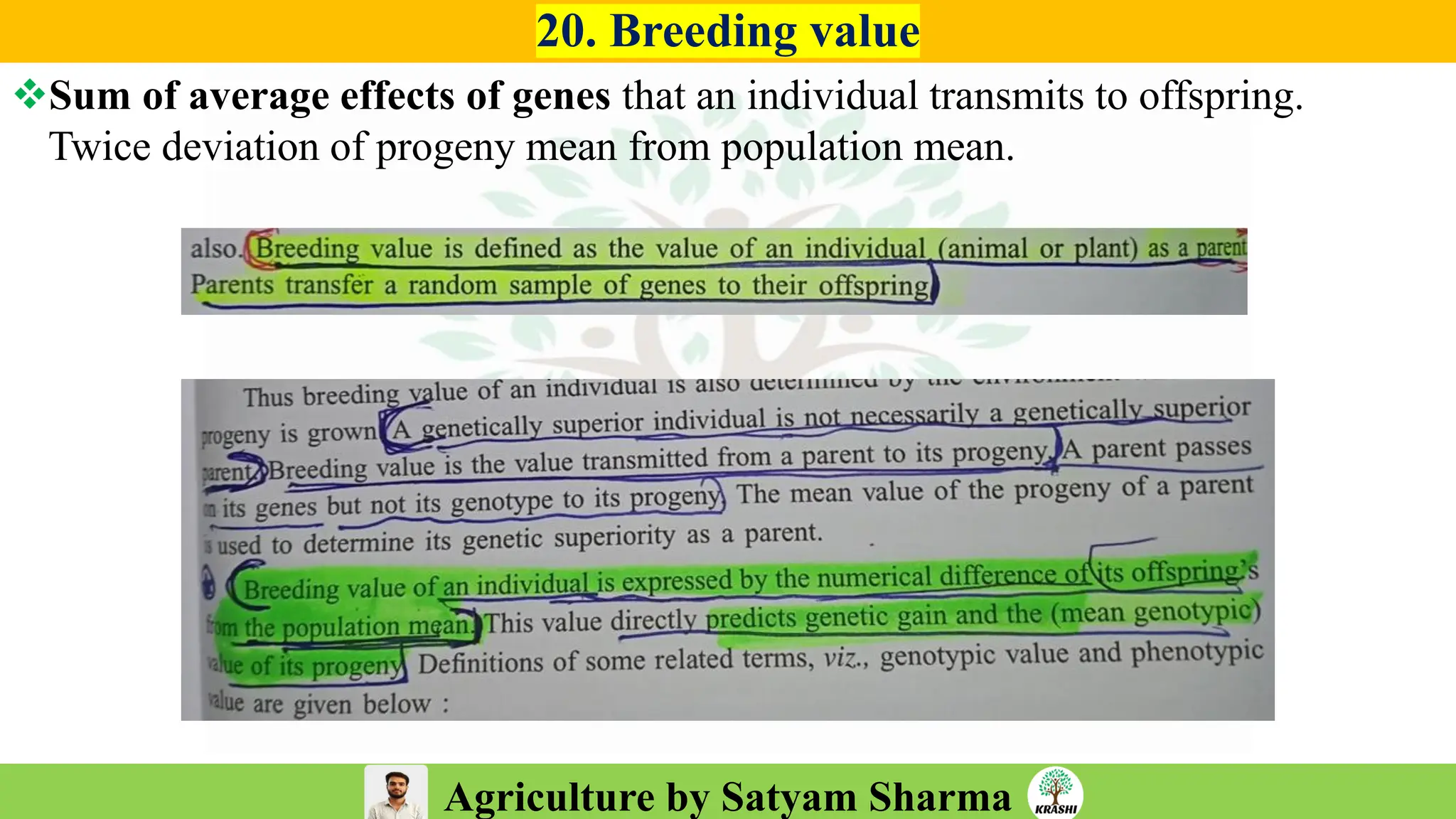
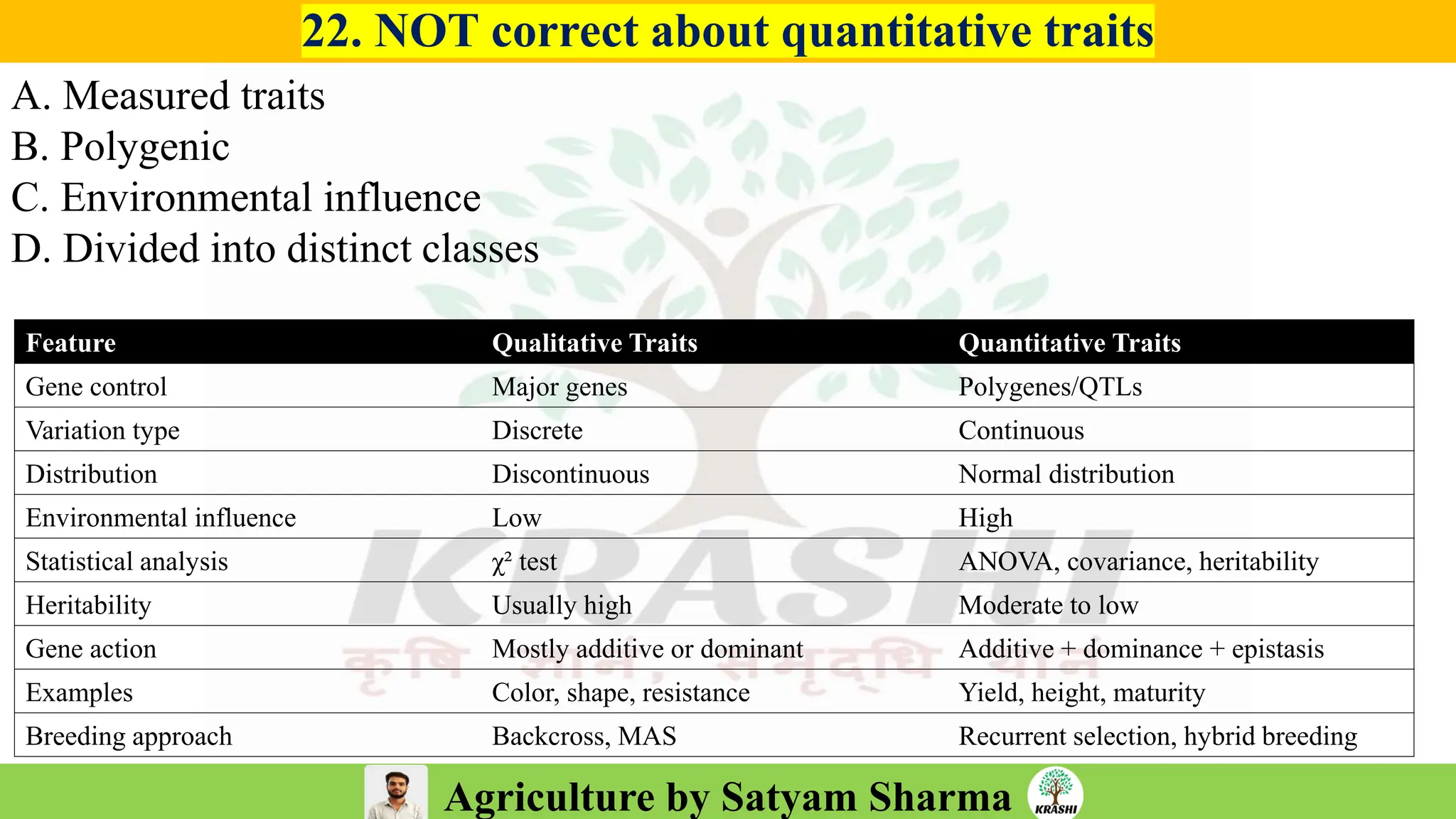
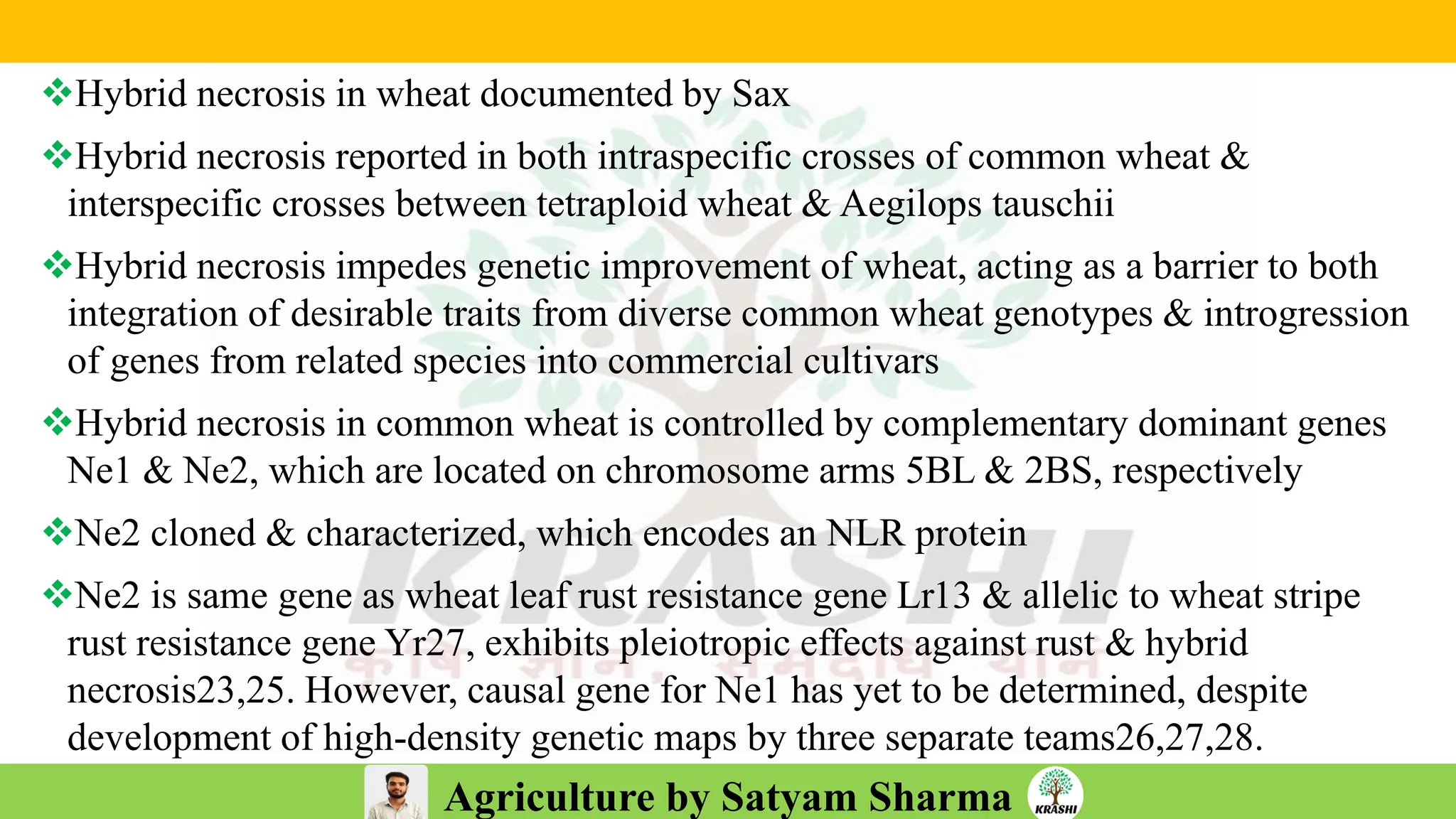
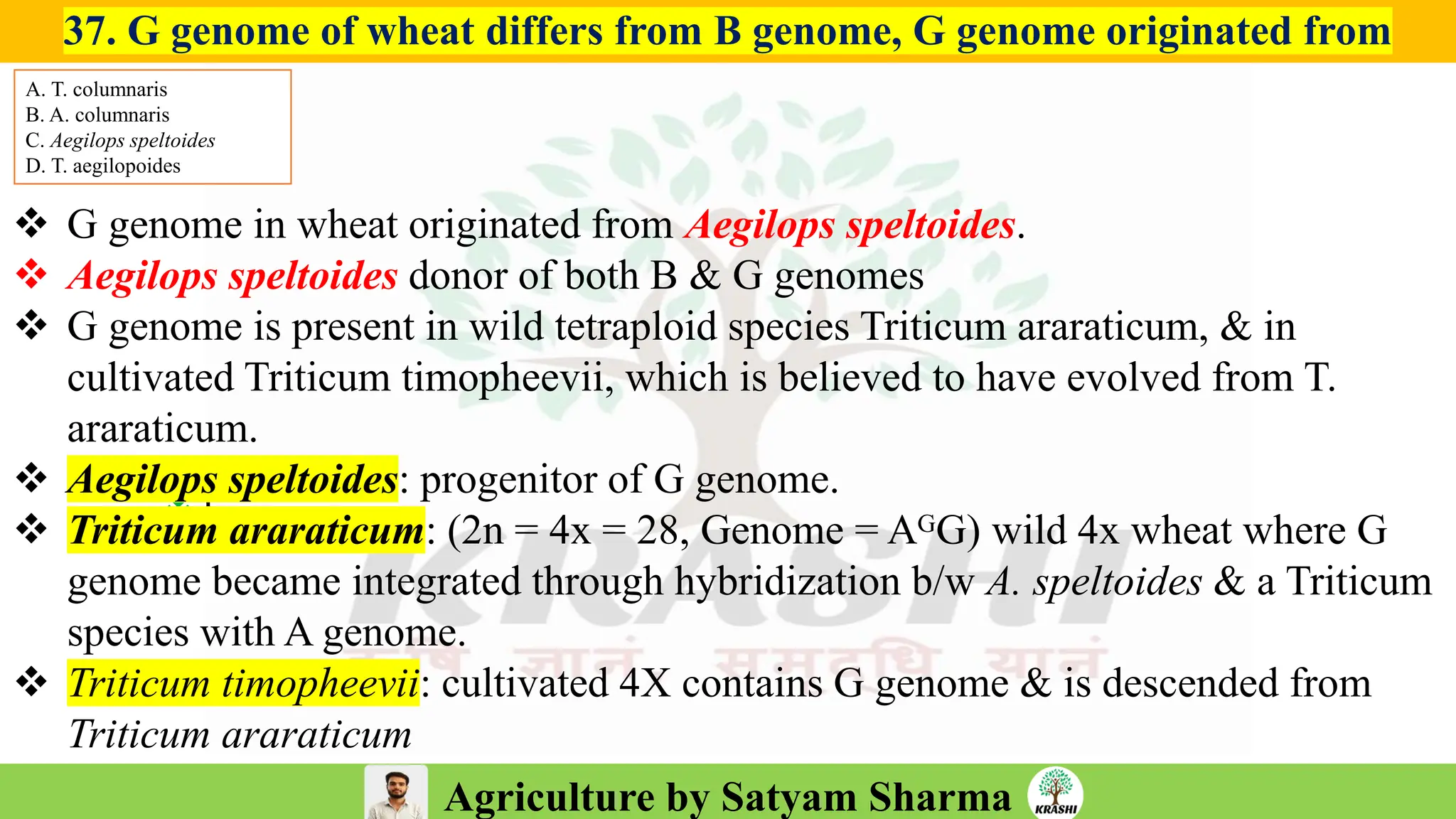
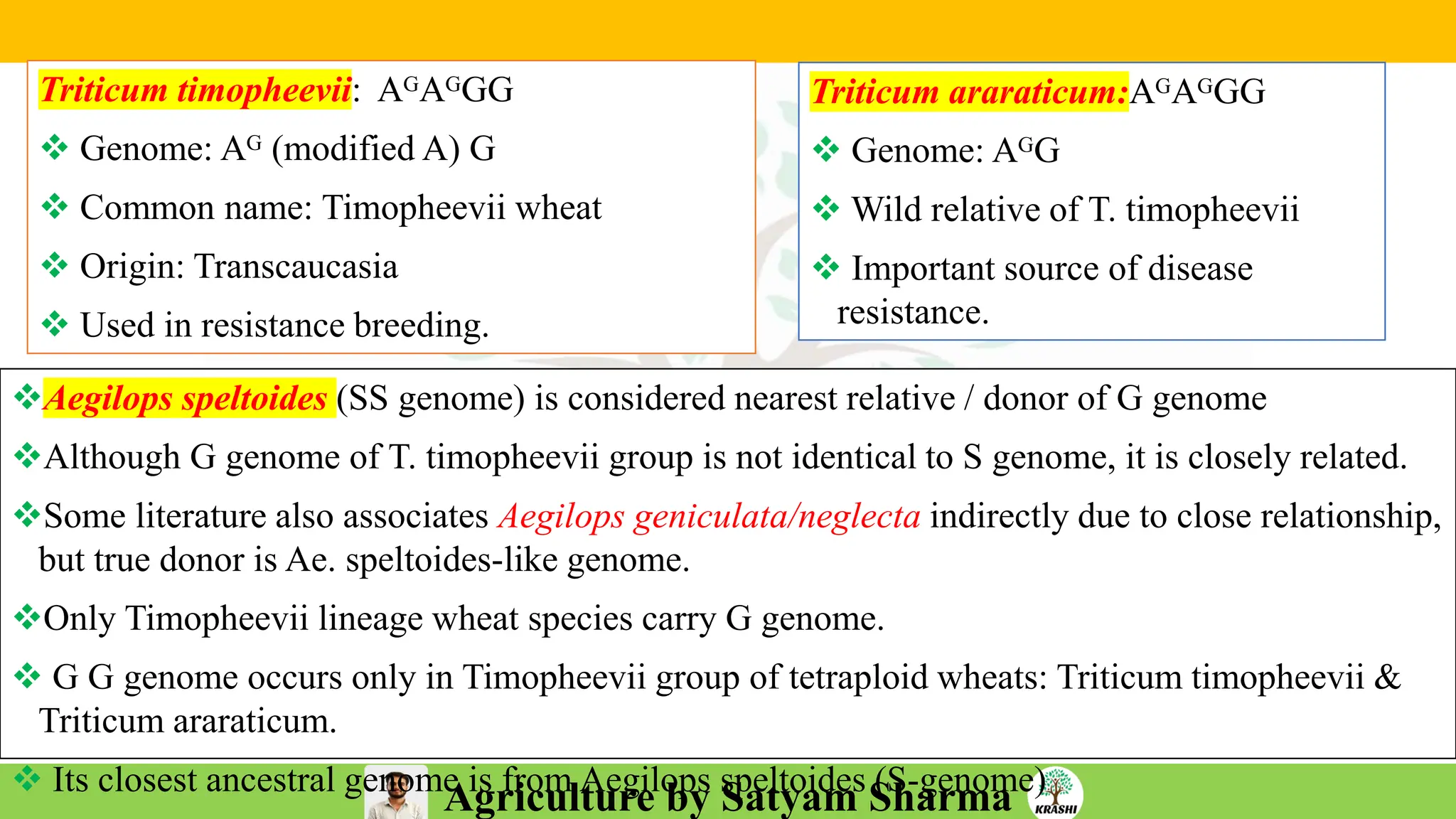
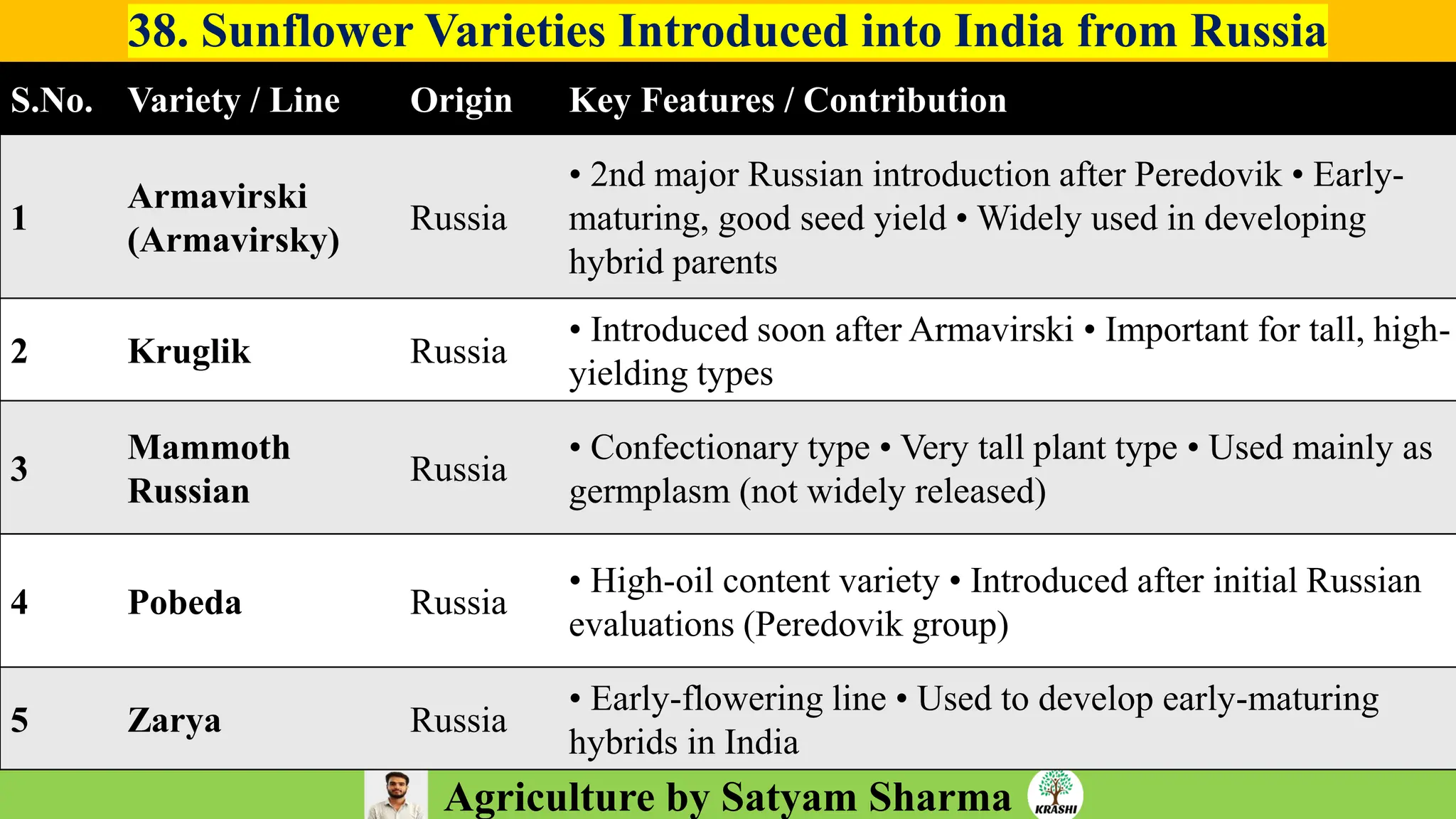
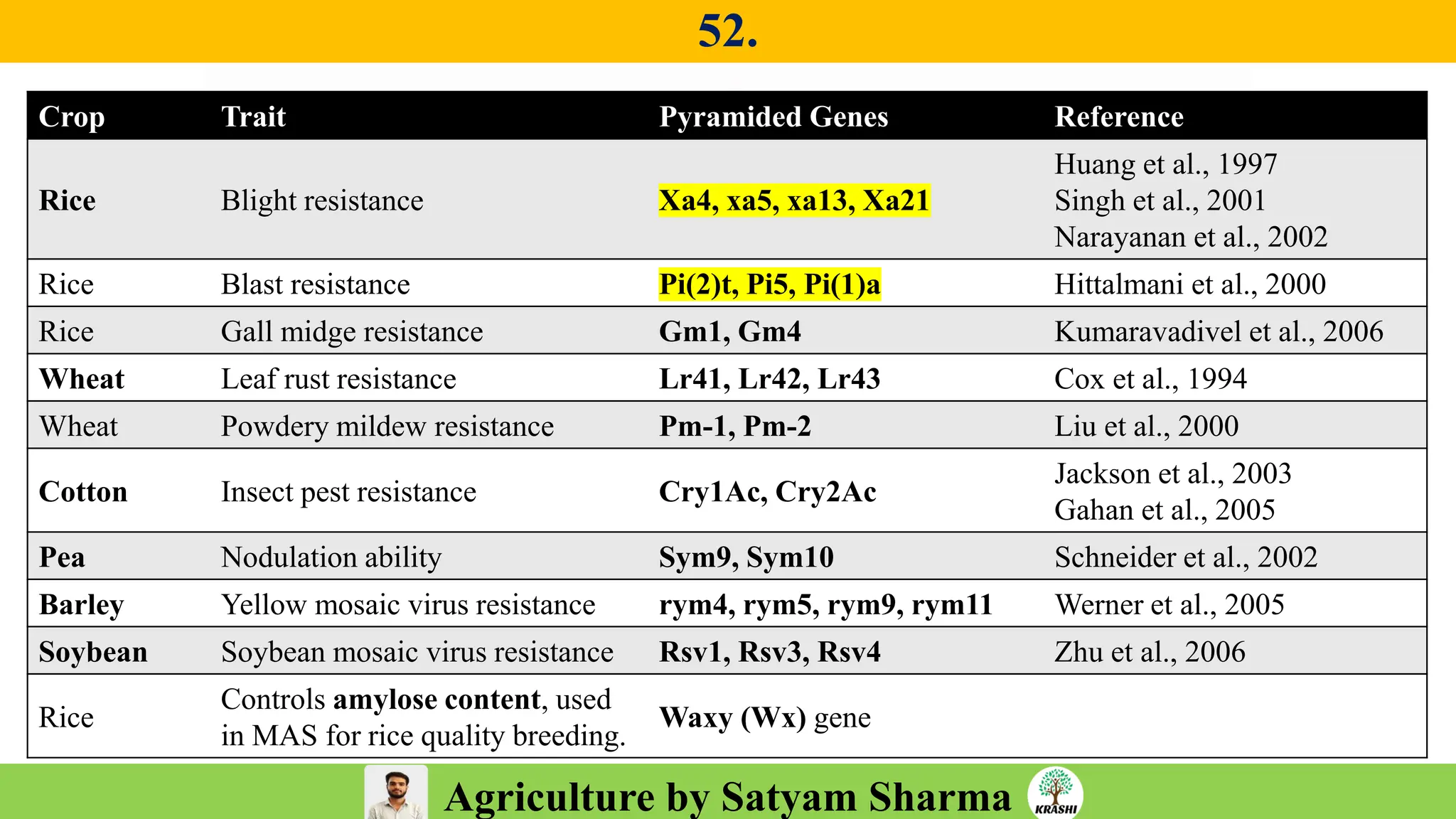
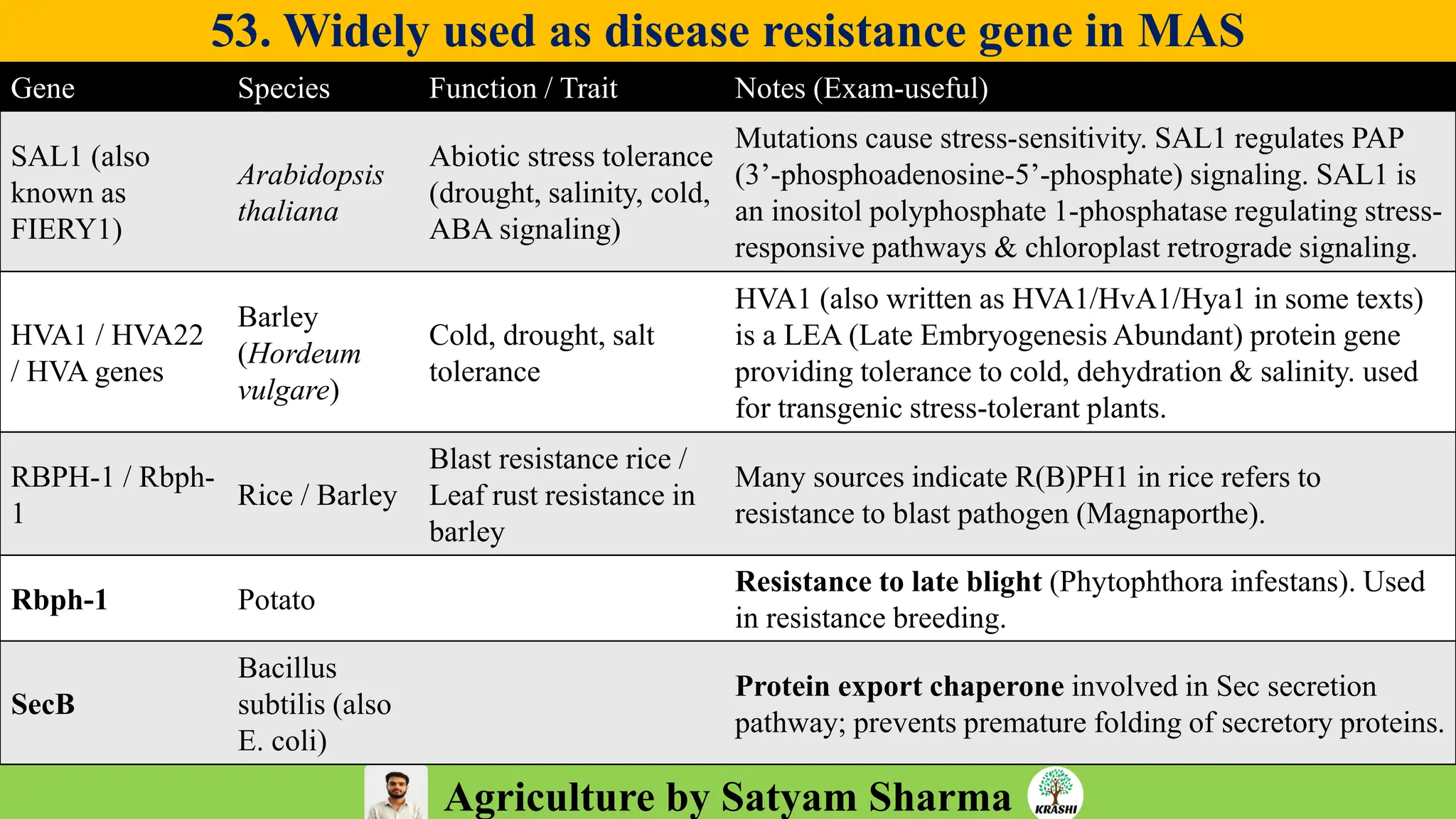
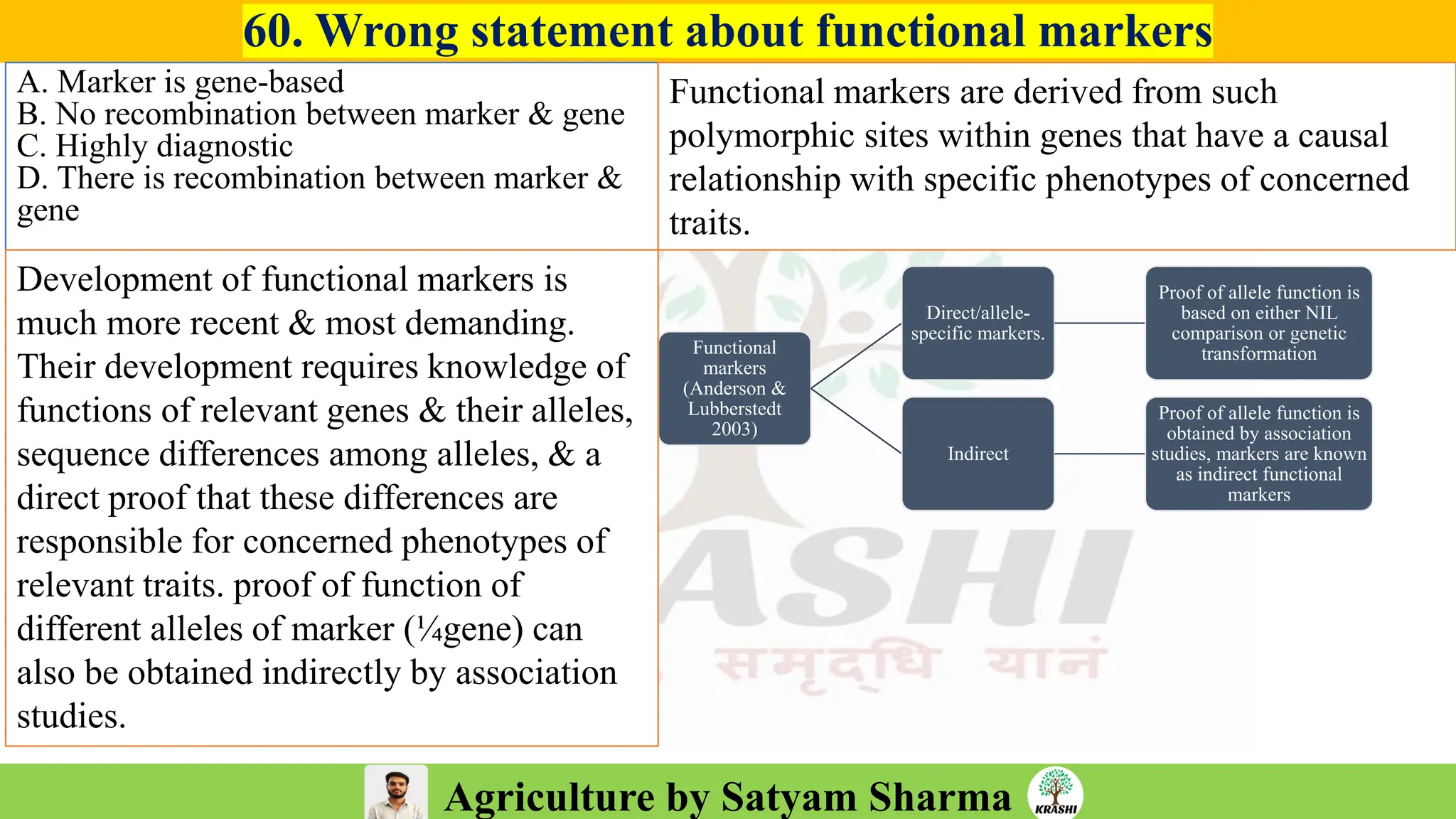
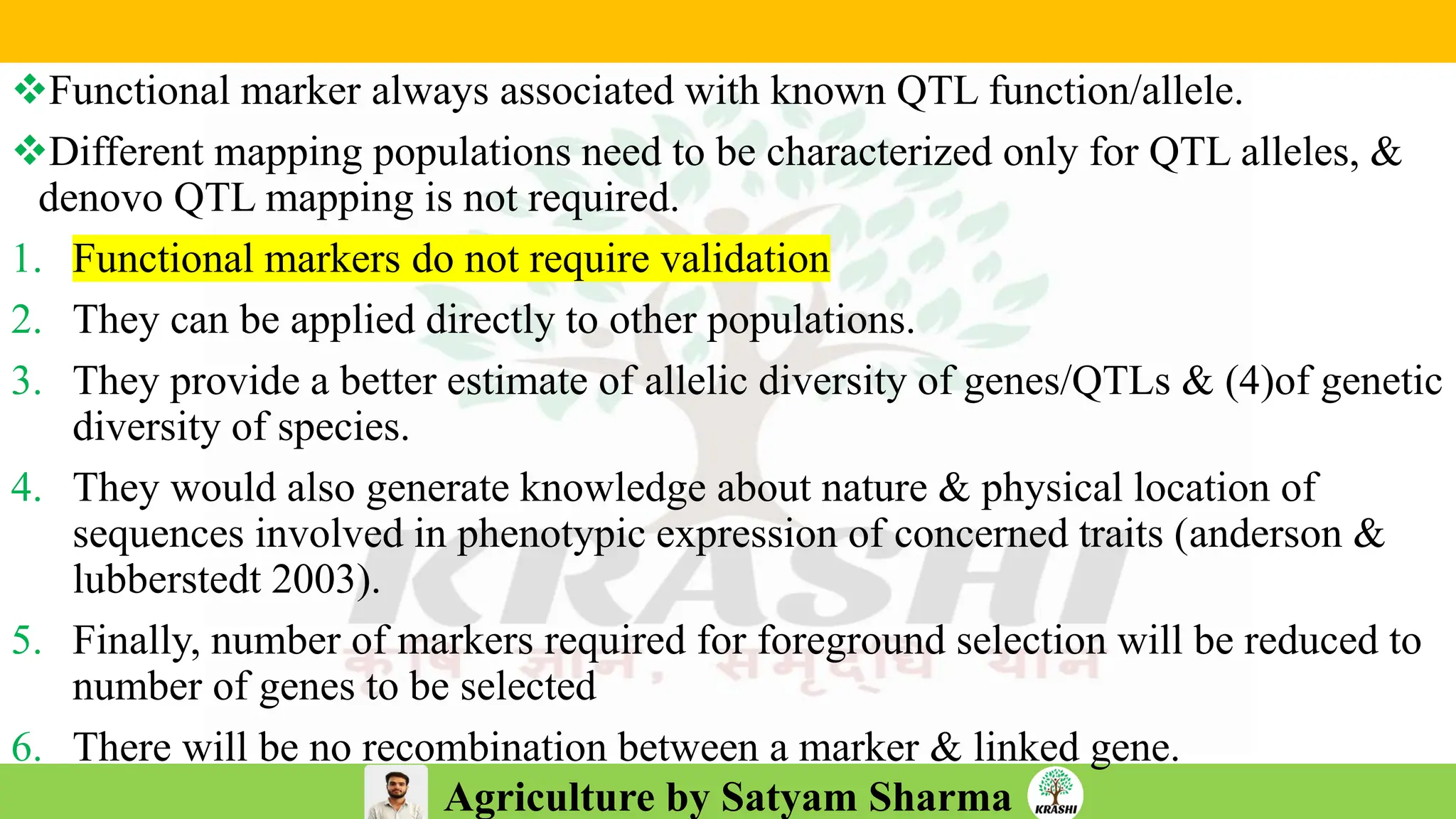
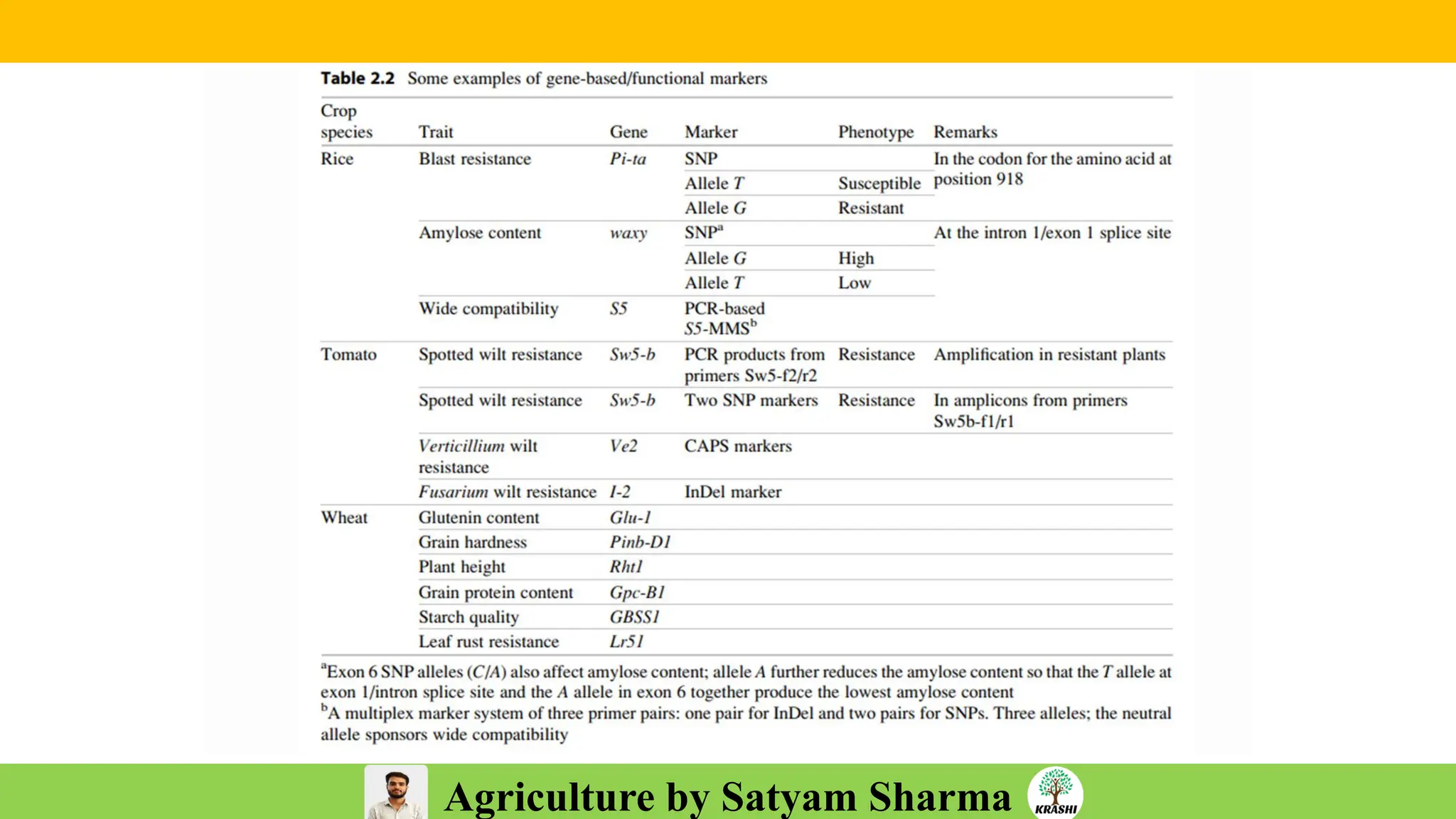
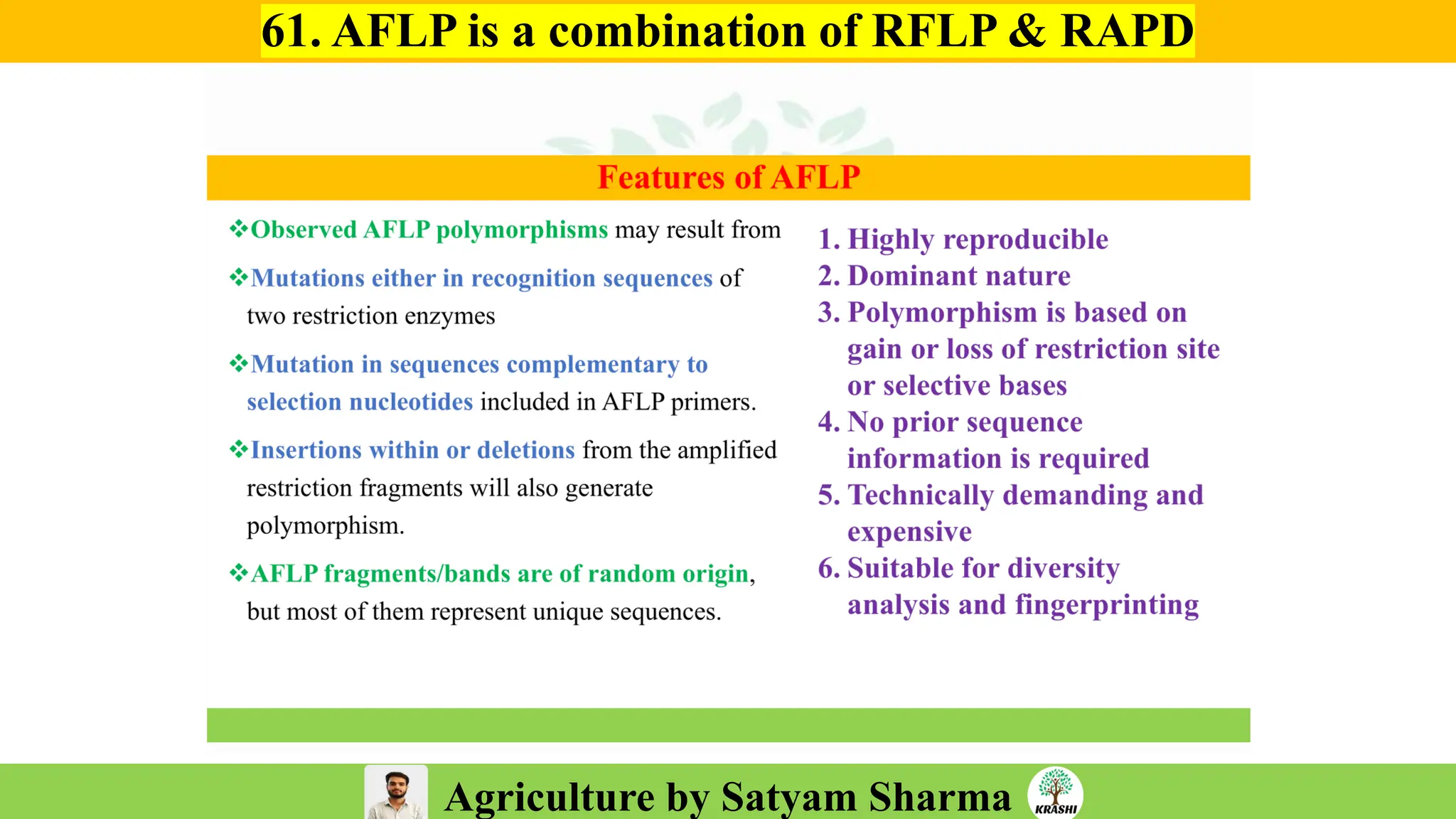
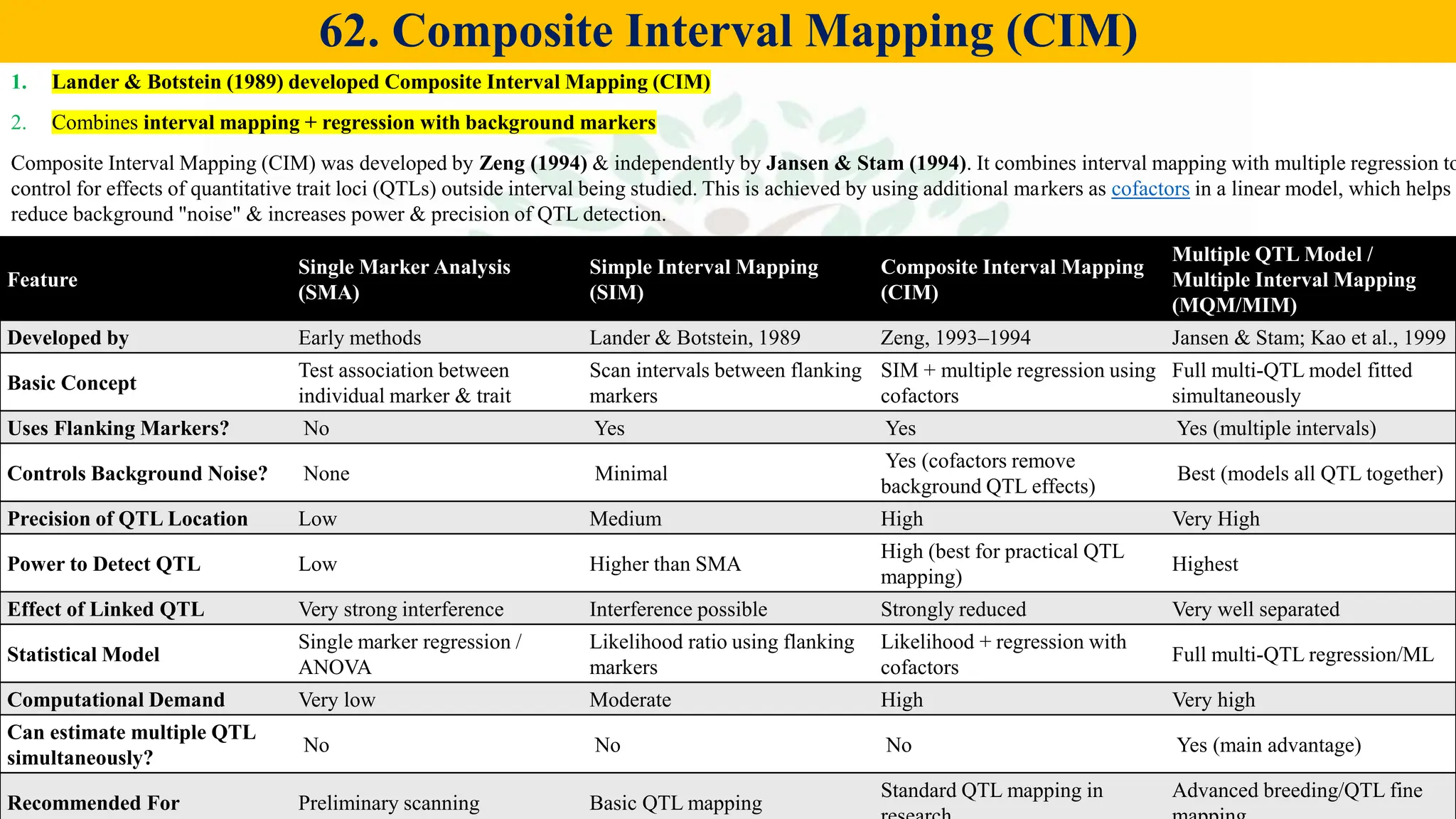
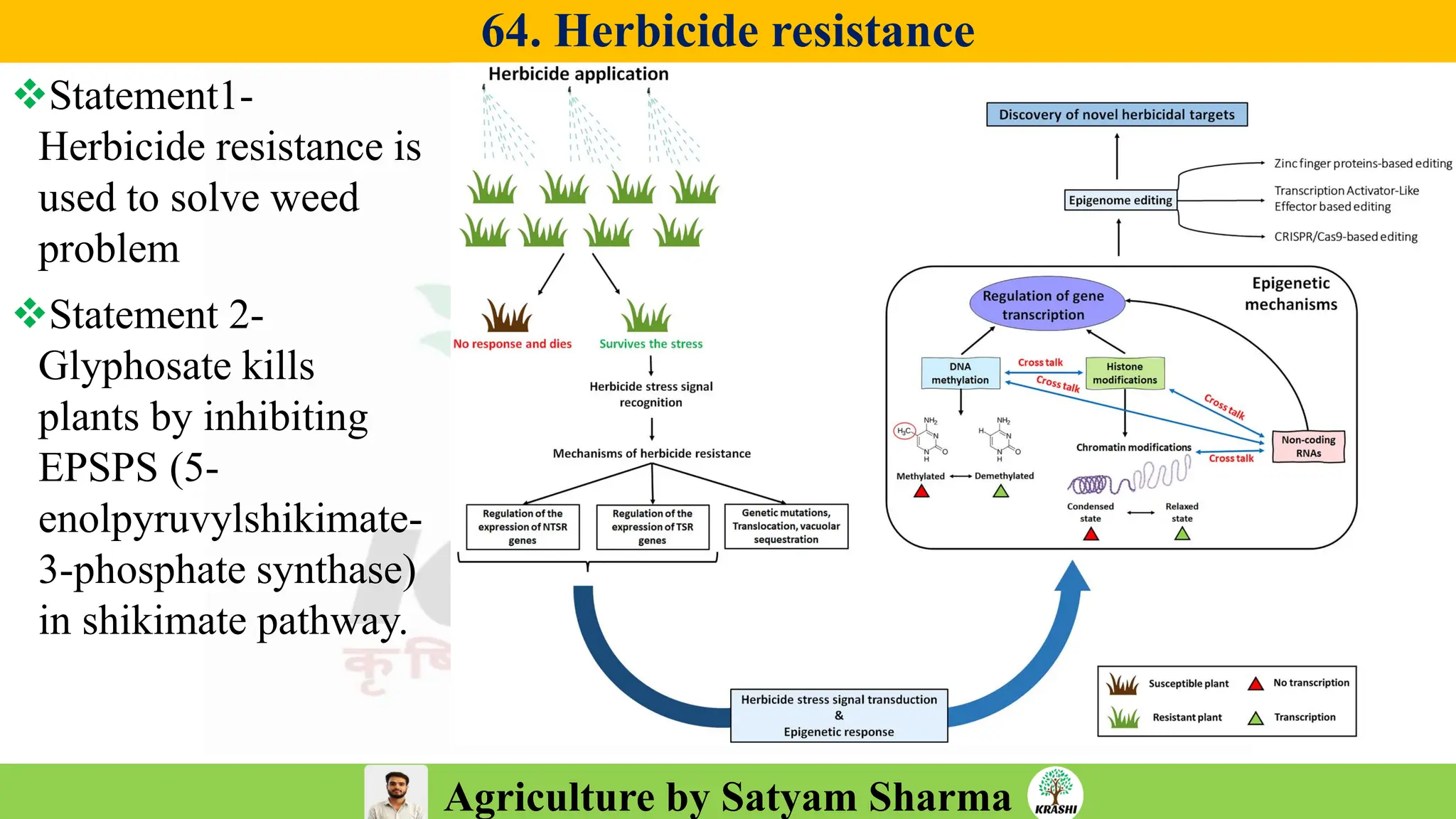
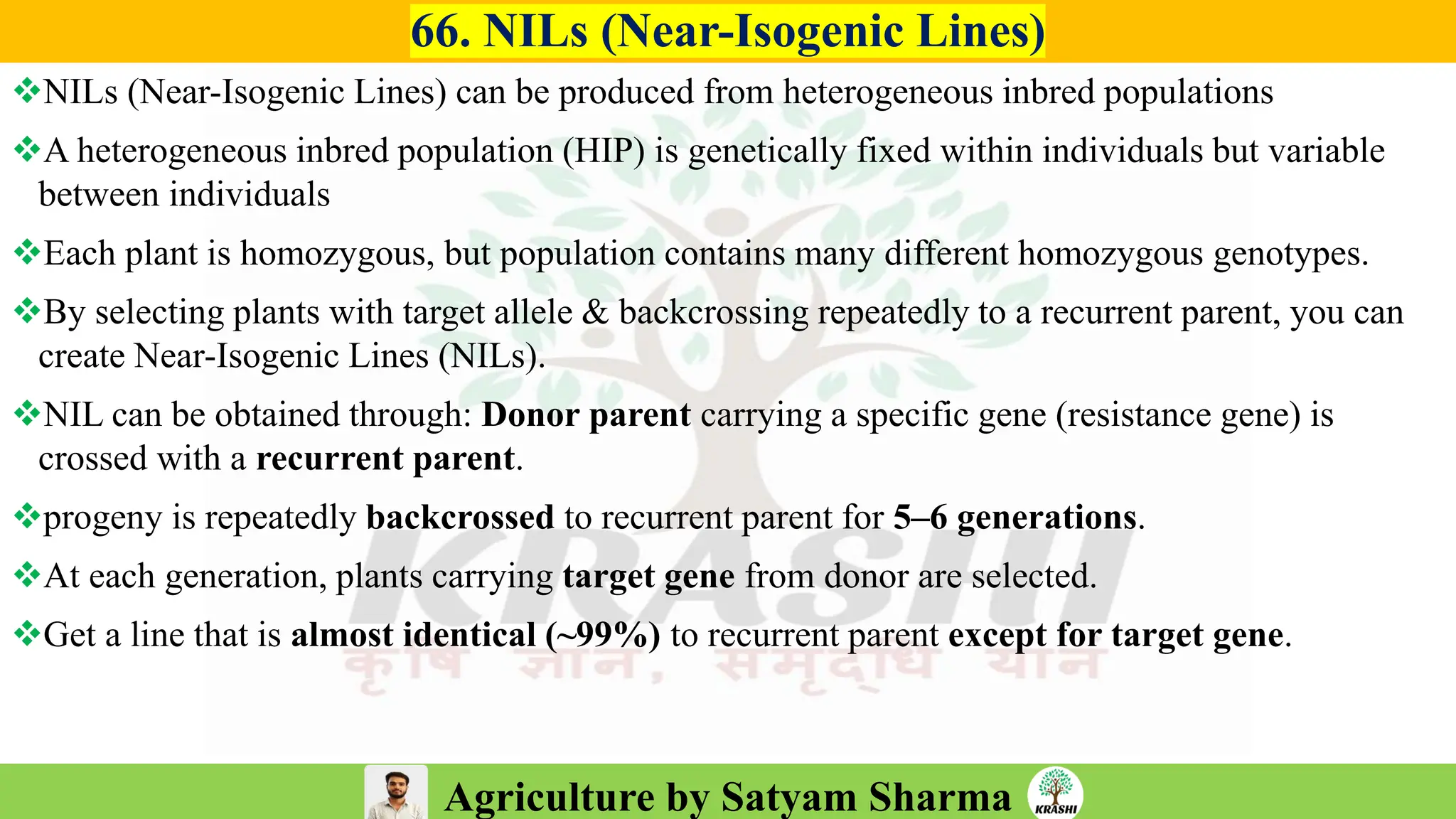
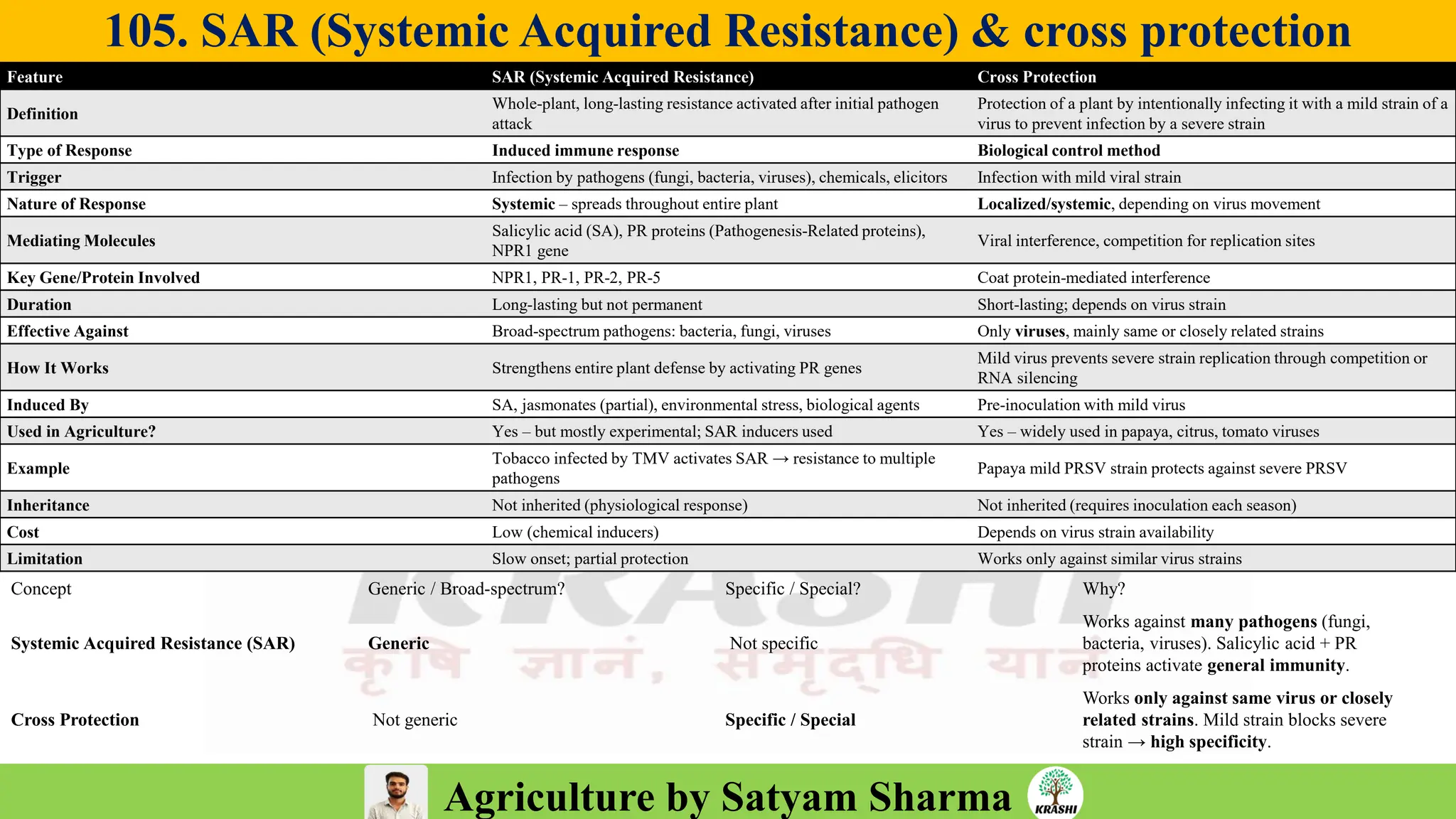
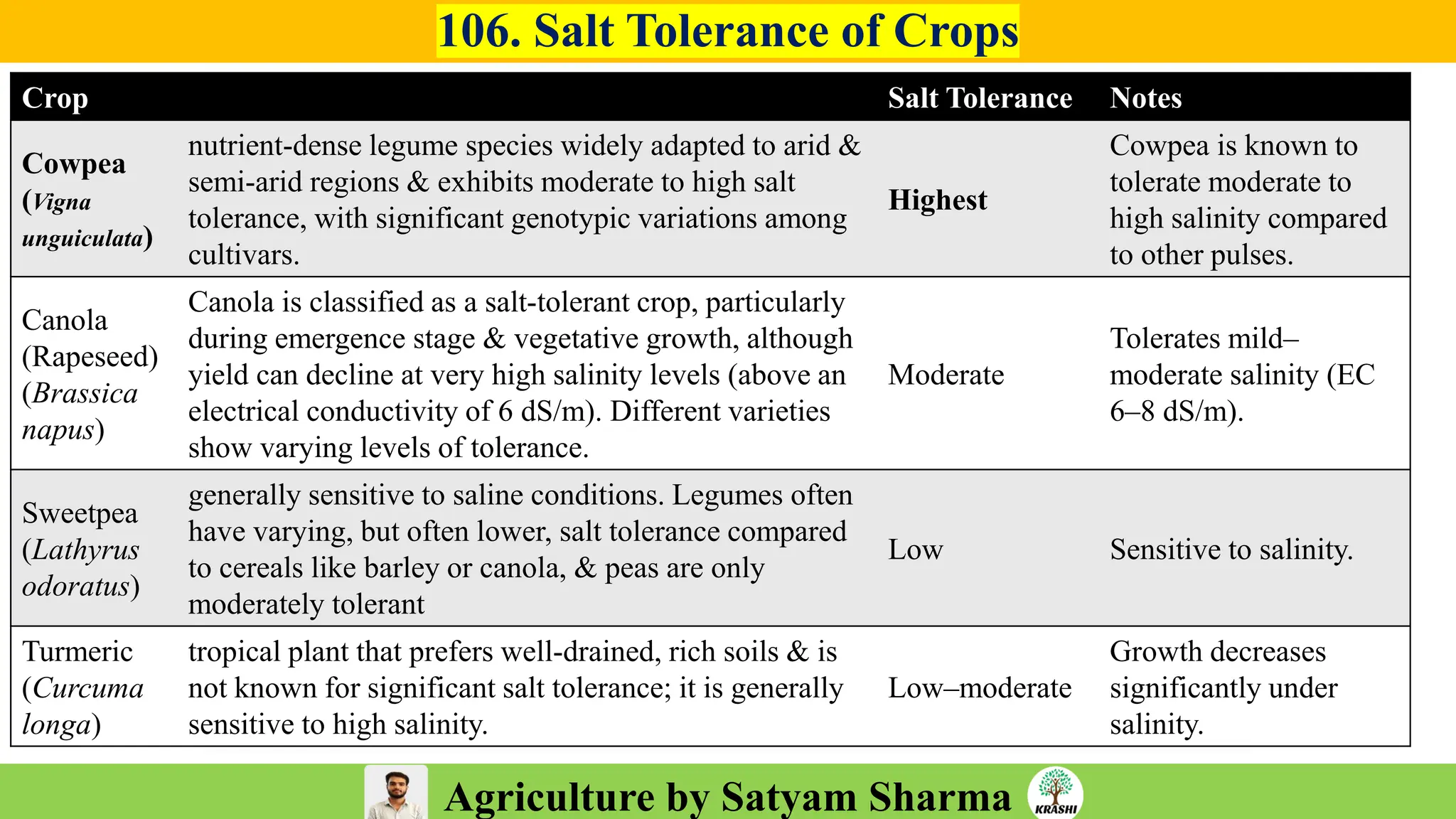
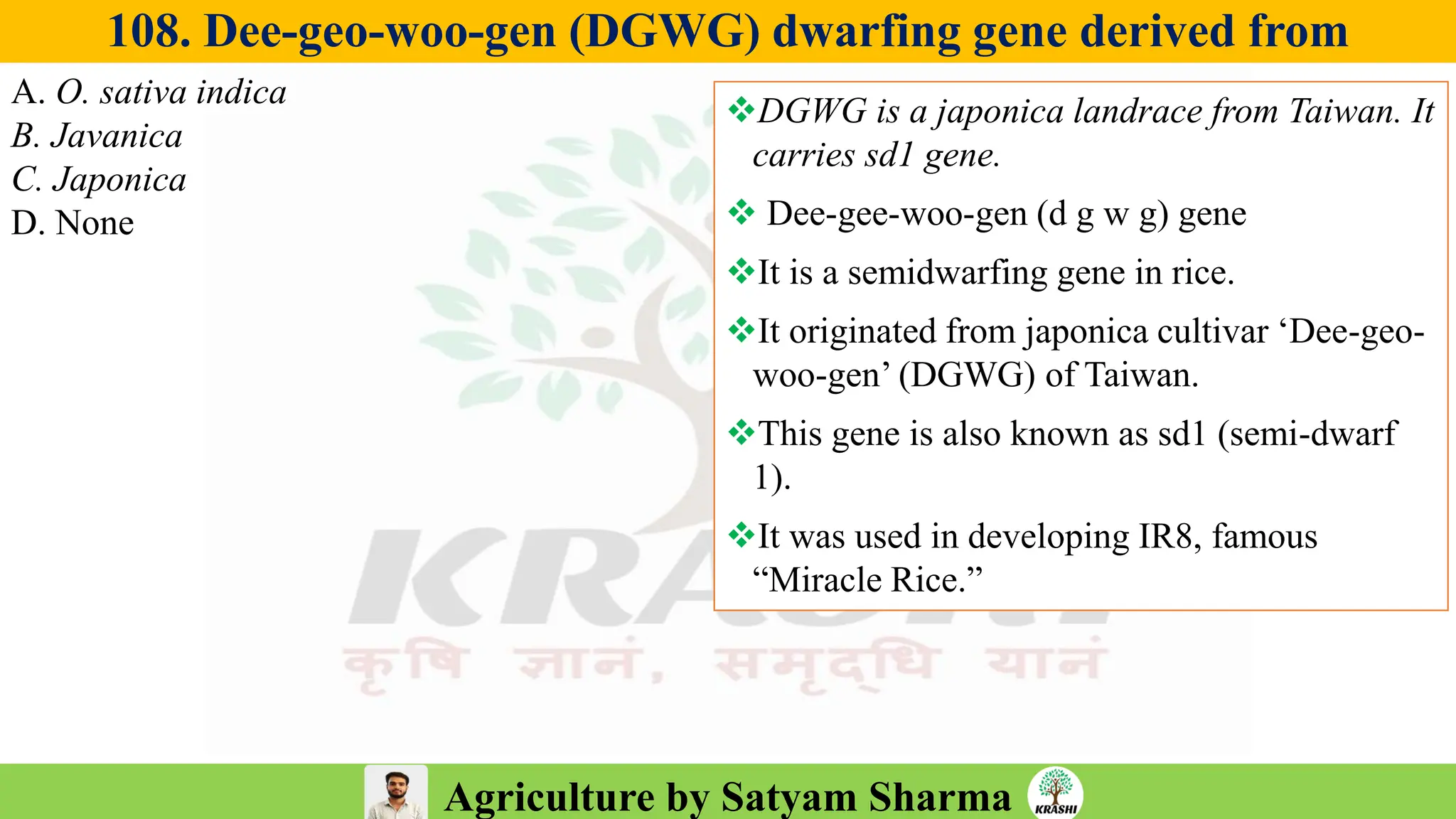
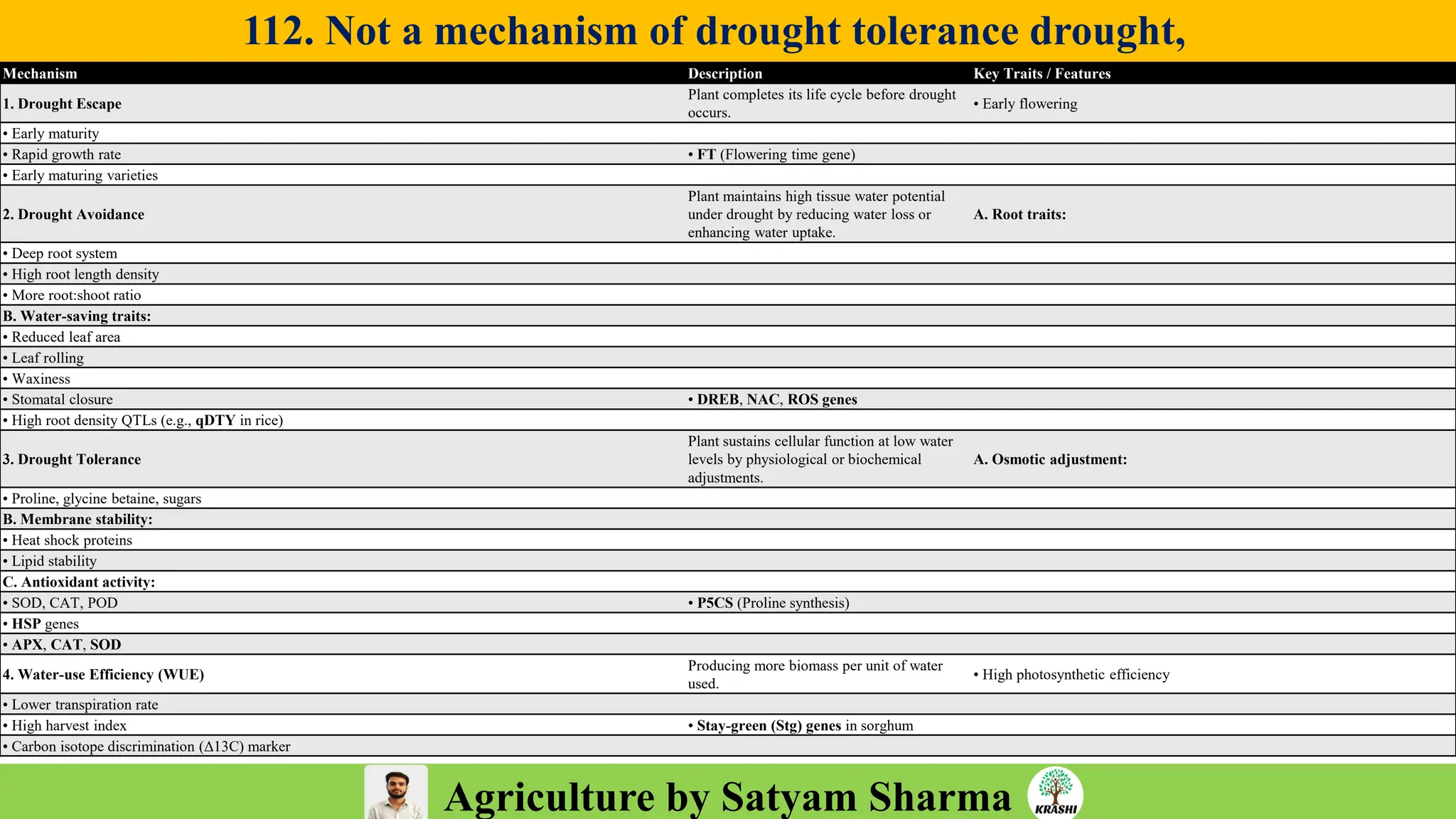
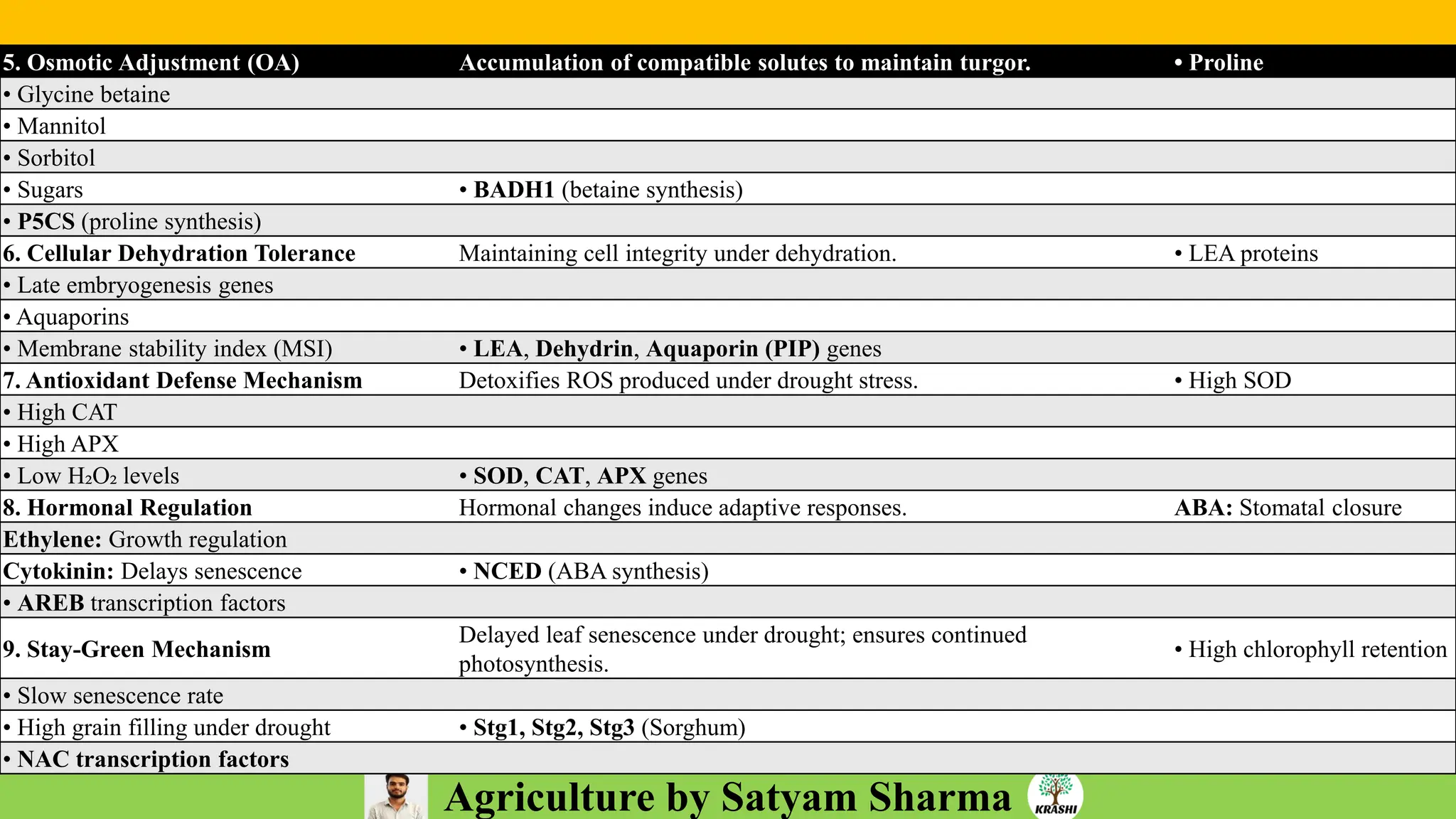
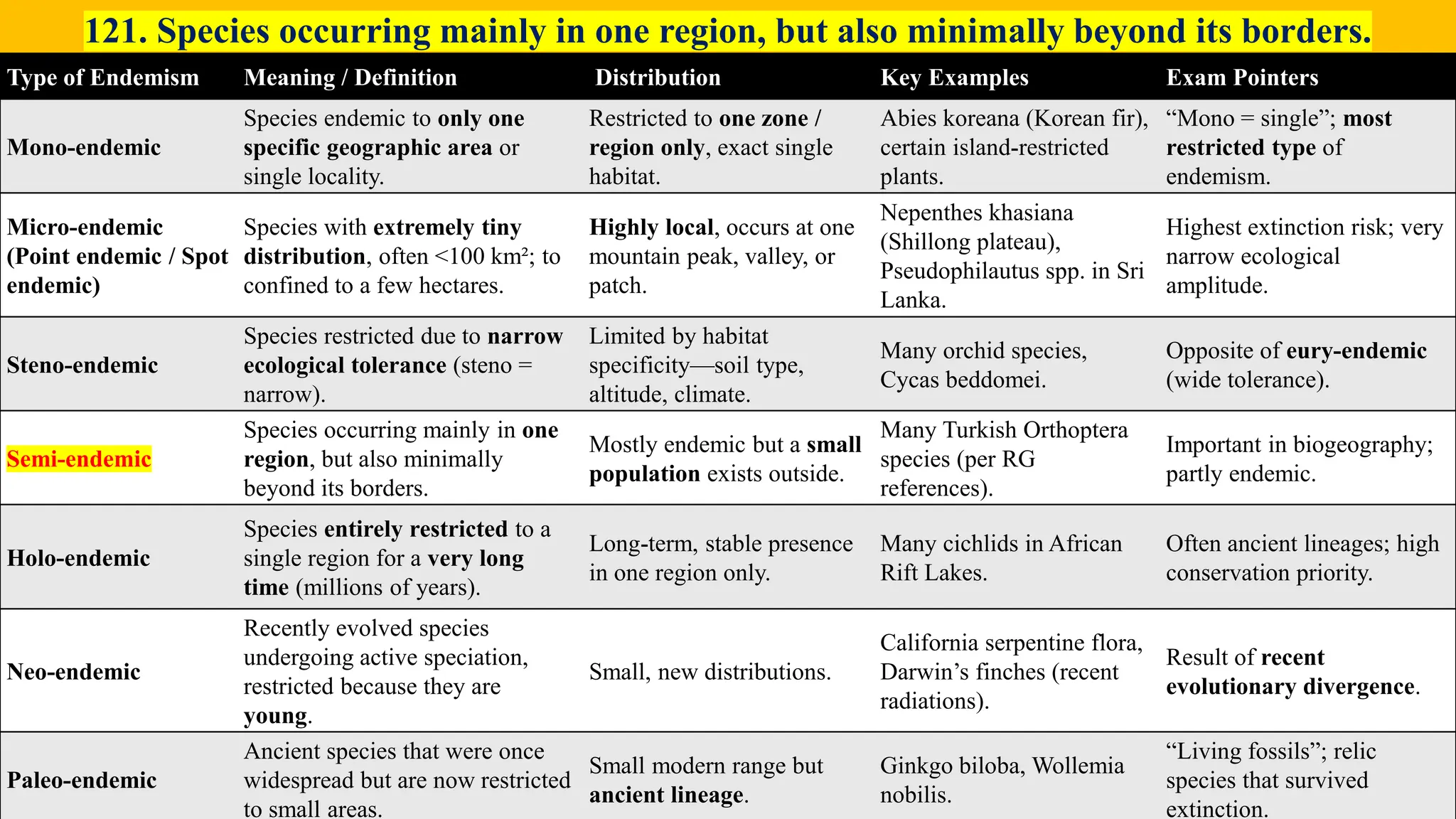
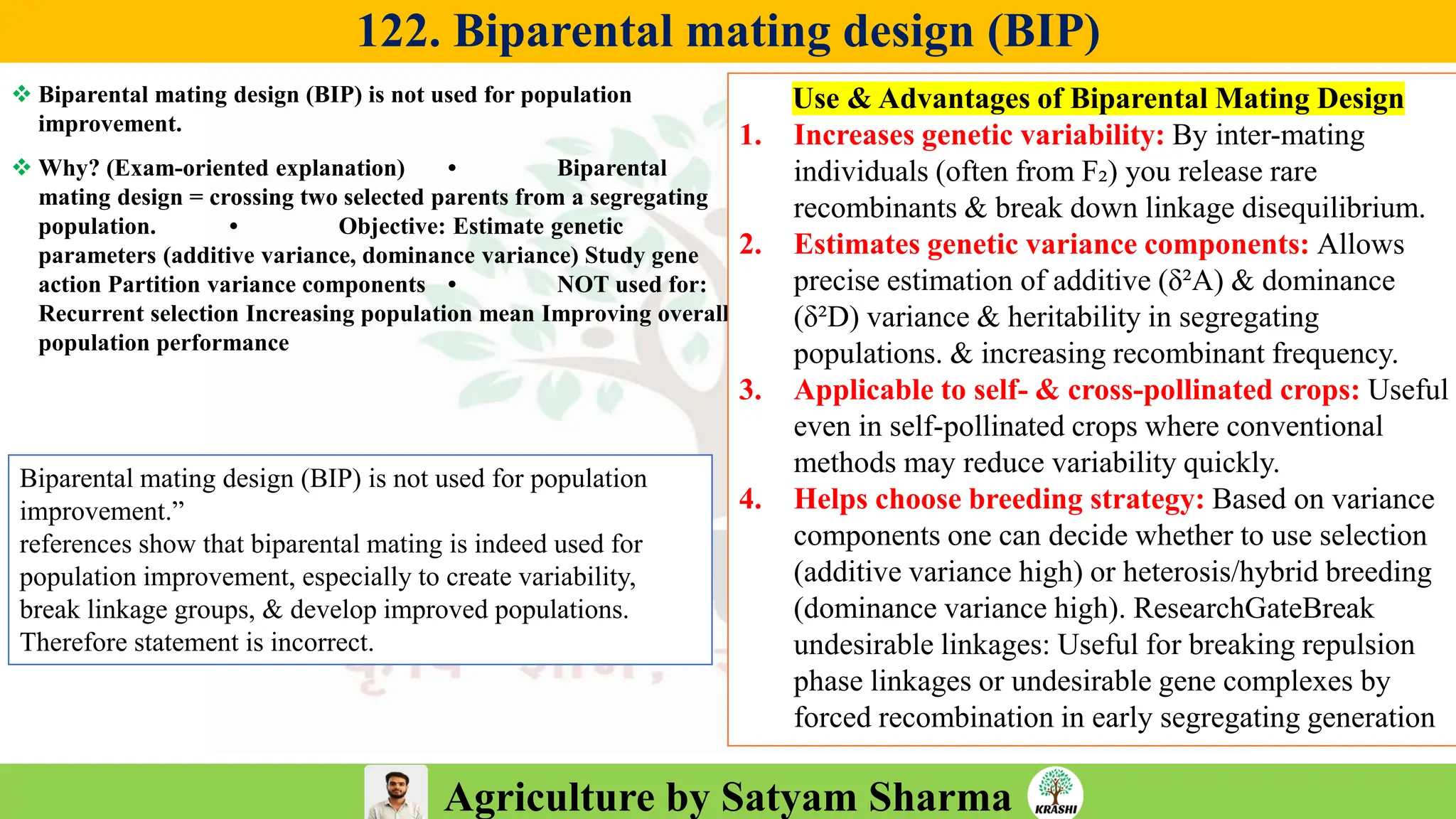
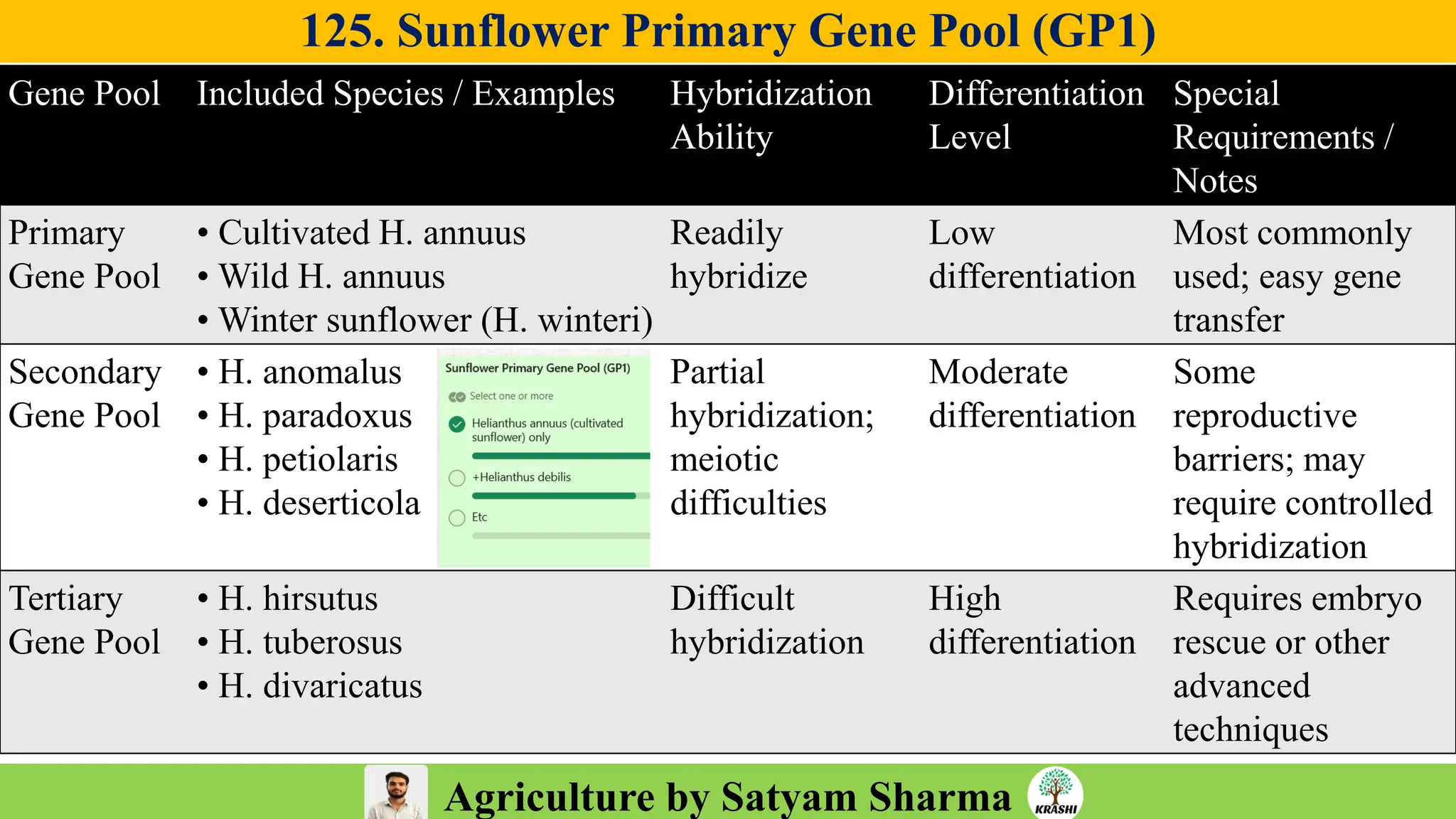
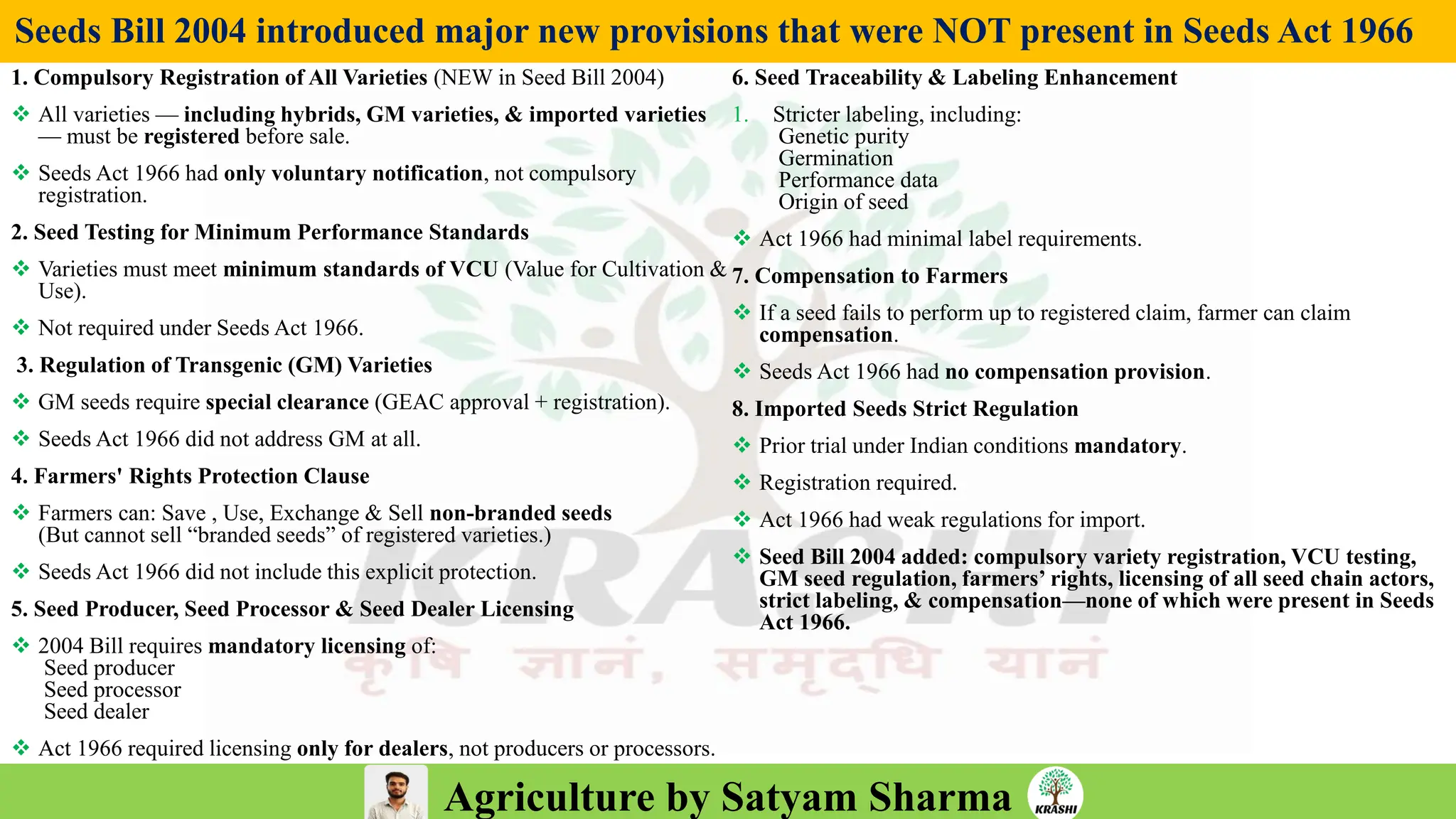
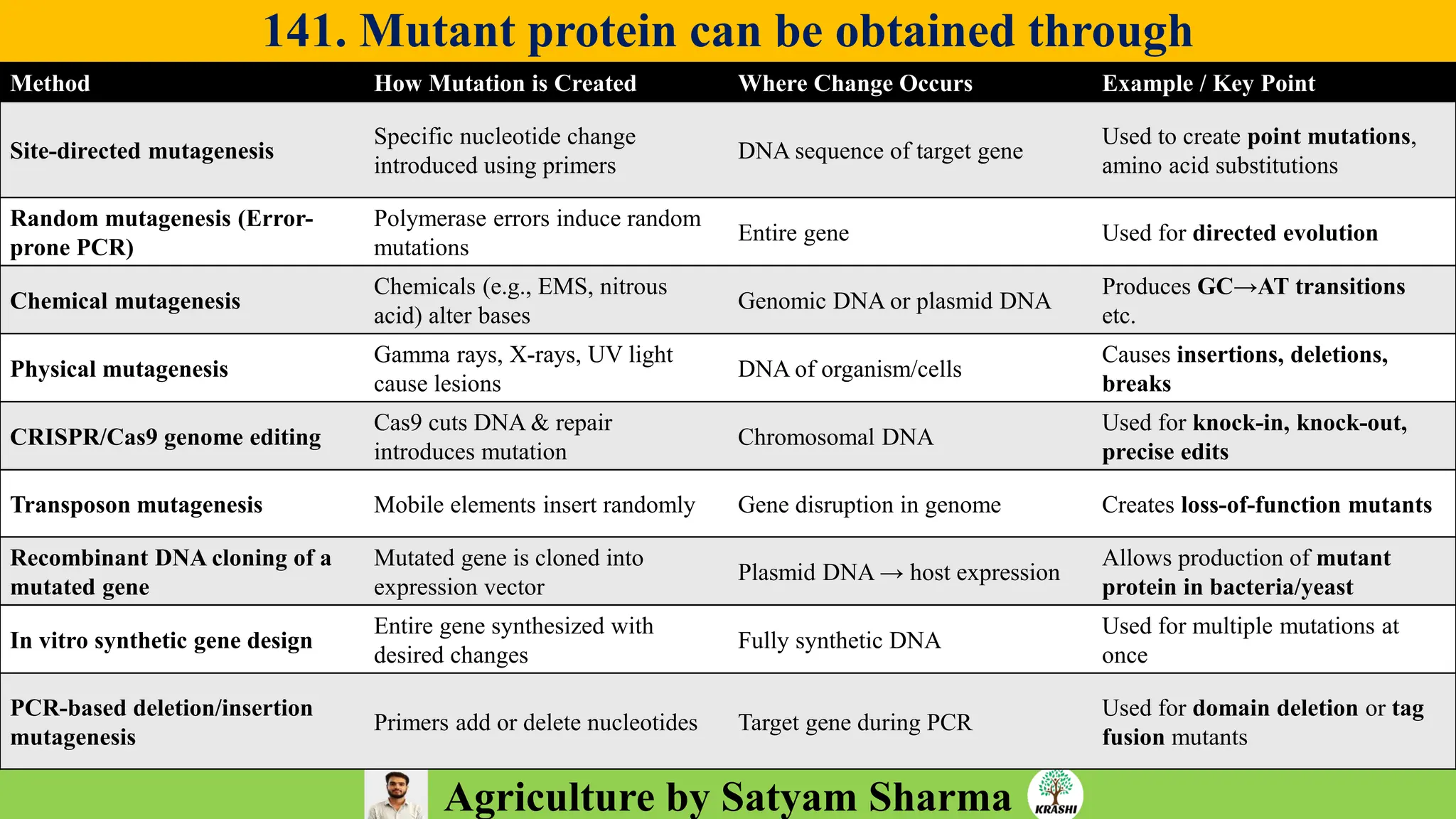
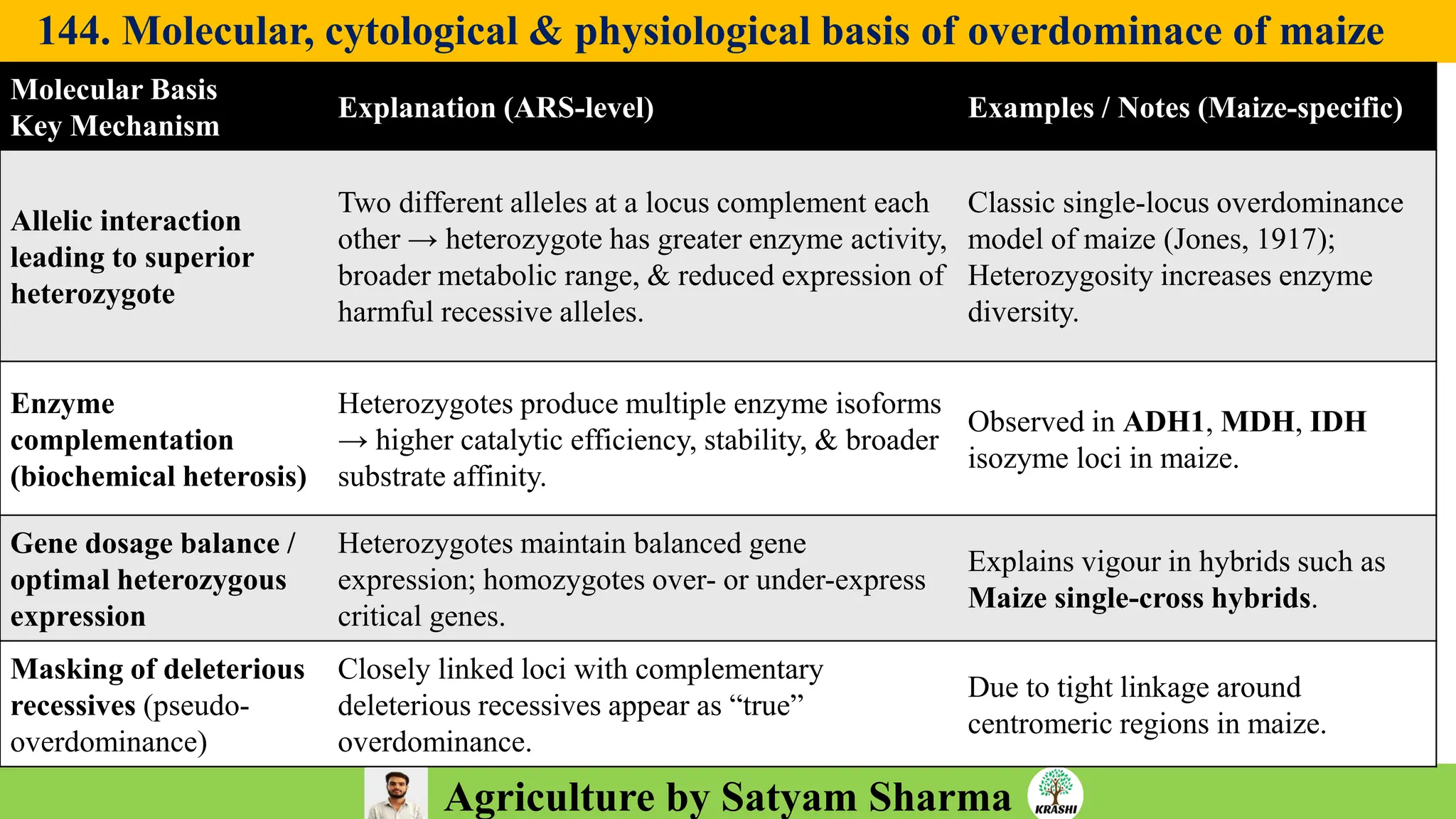
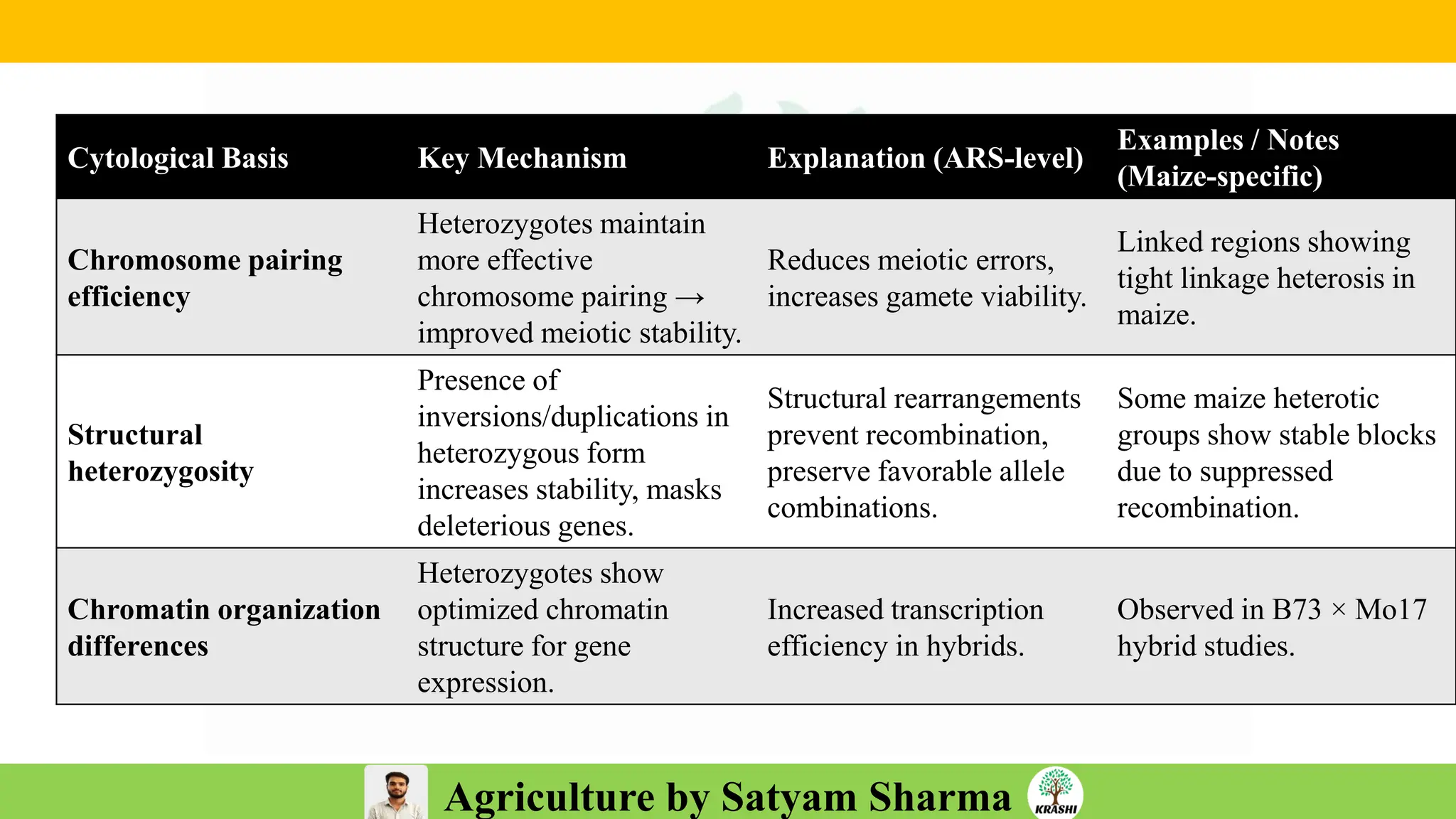
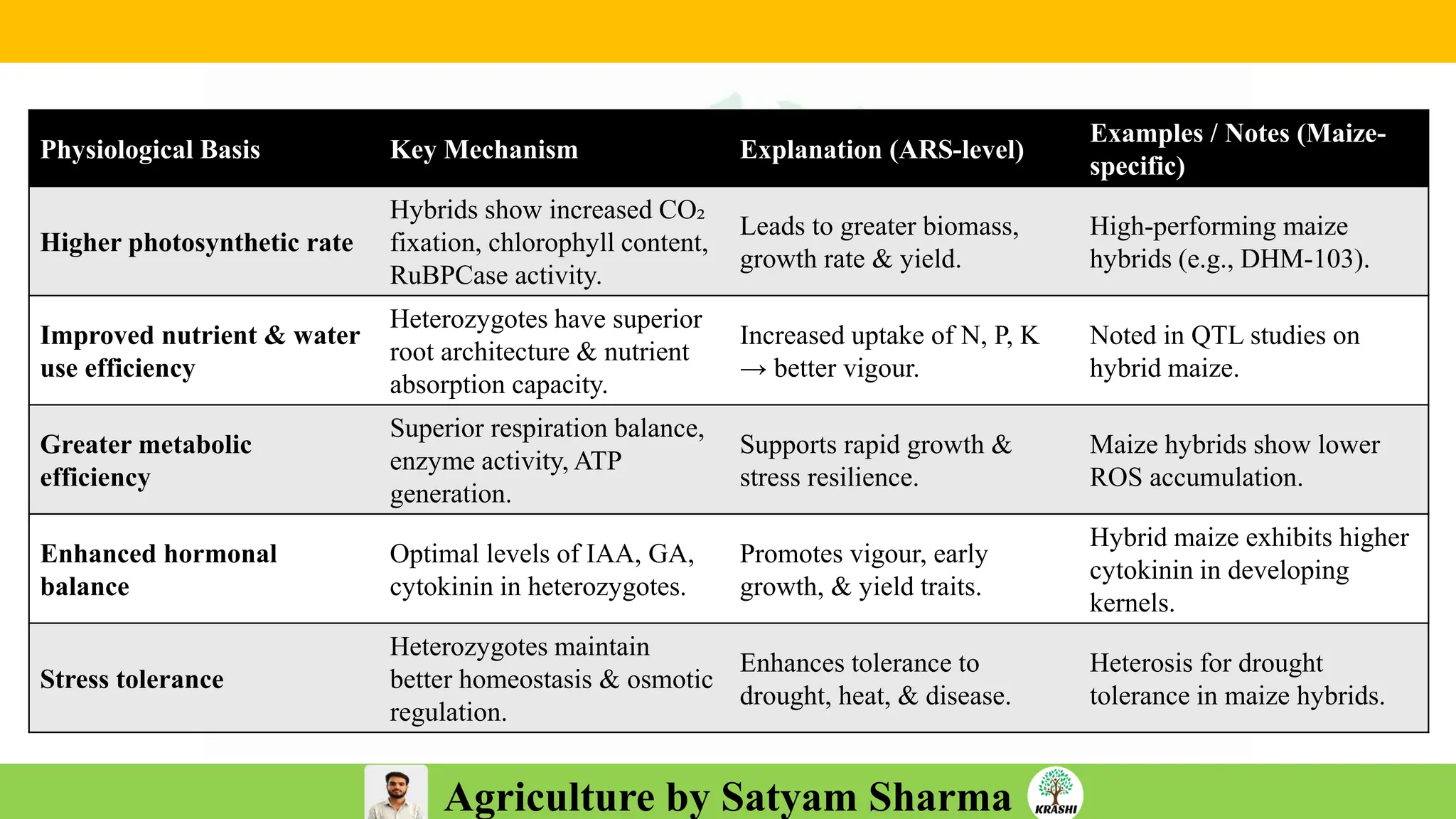
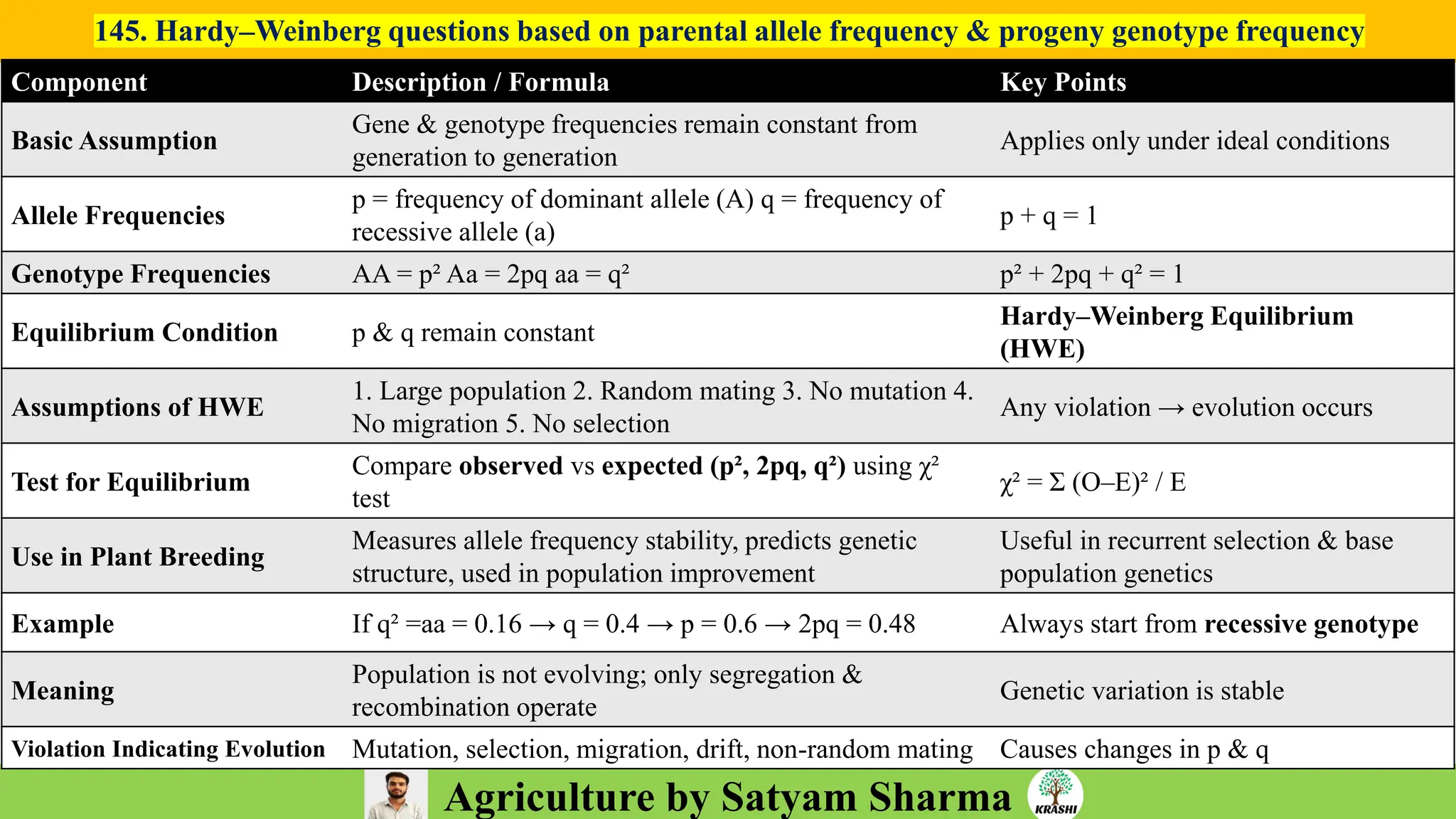
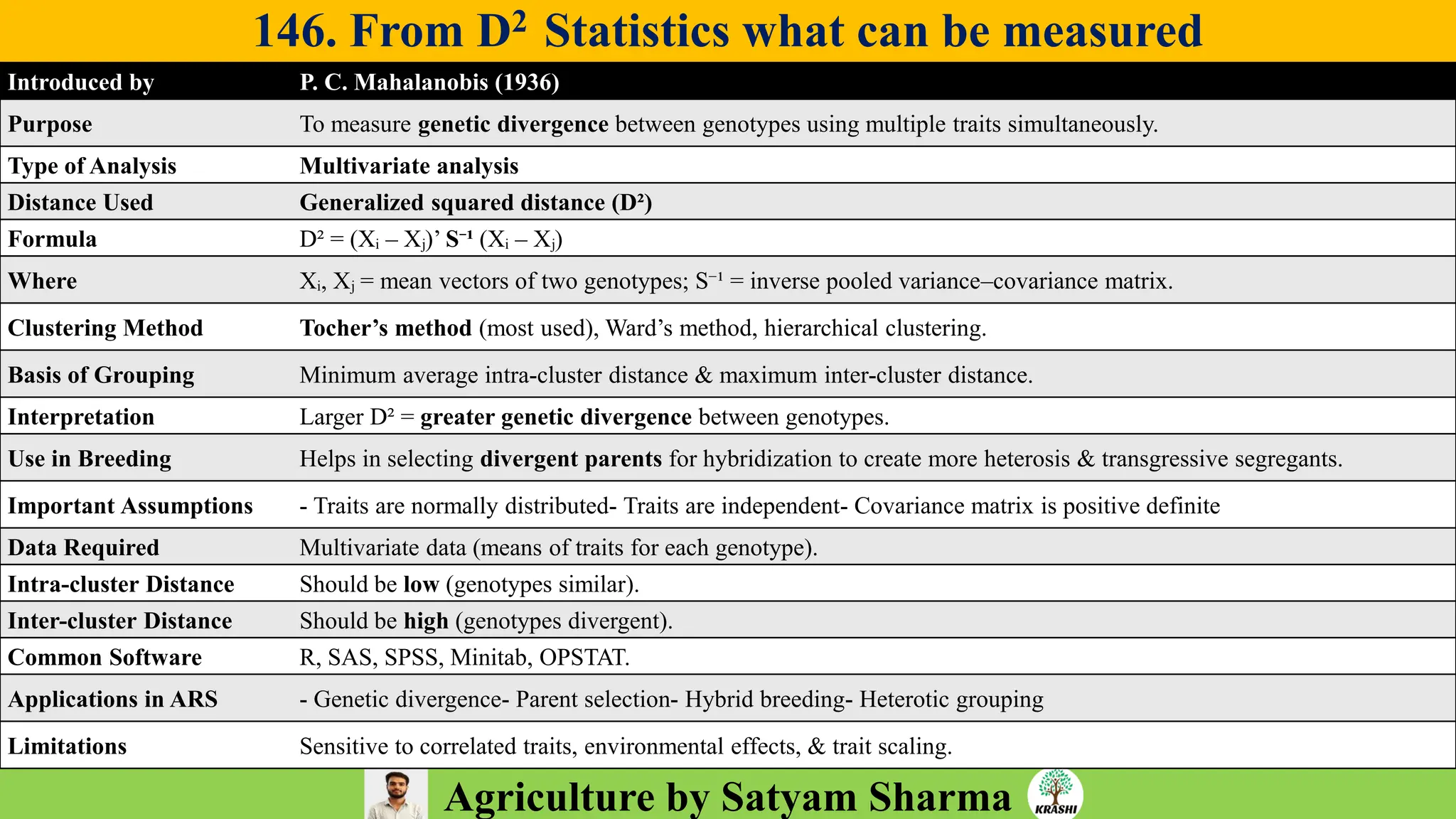
– Genetics (Epistasis, Gene action, QTL)
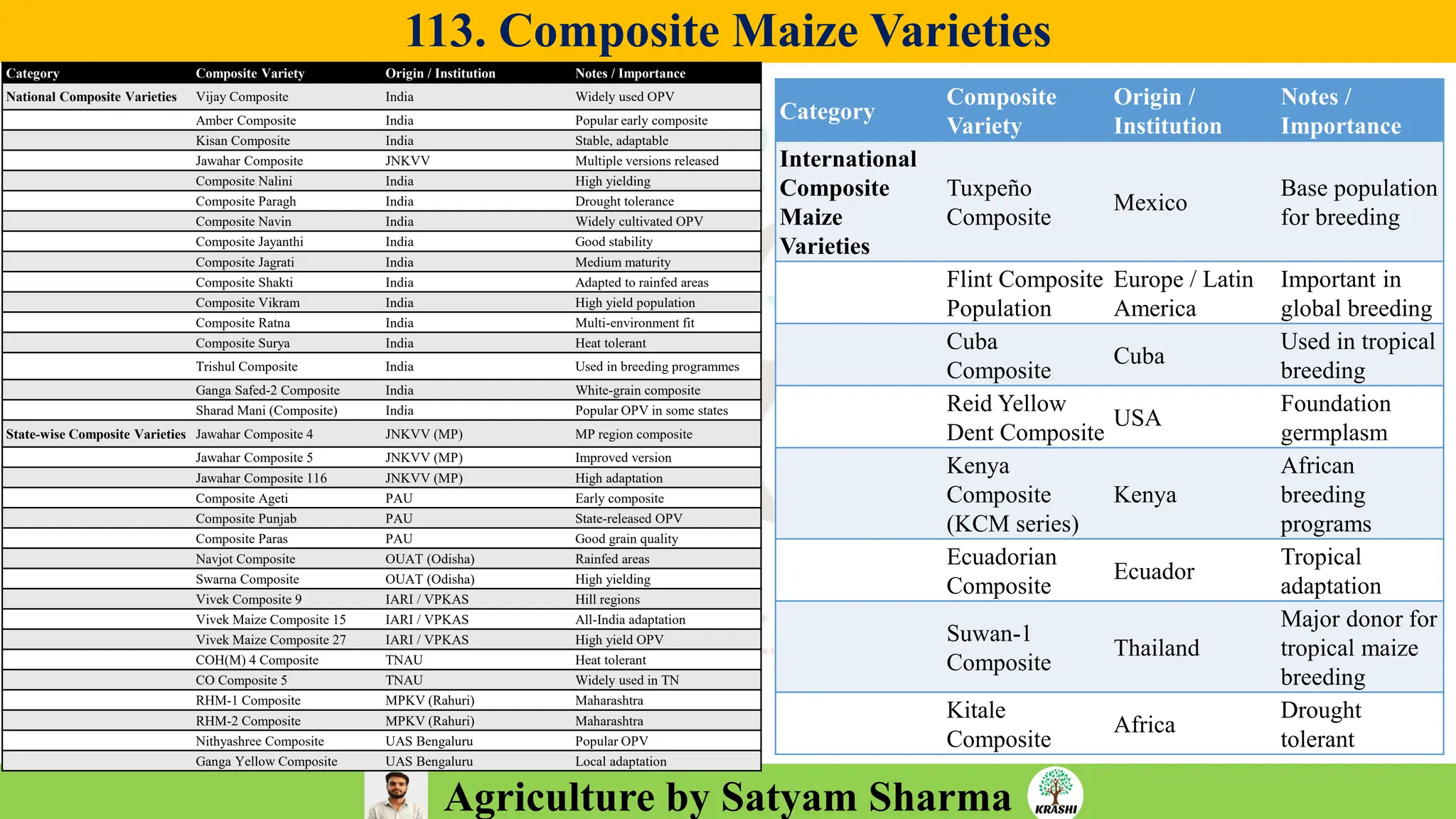
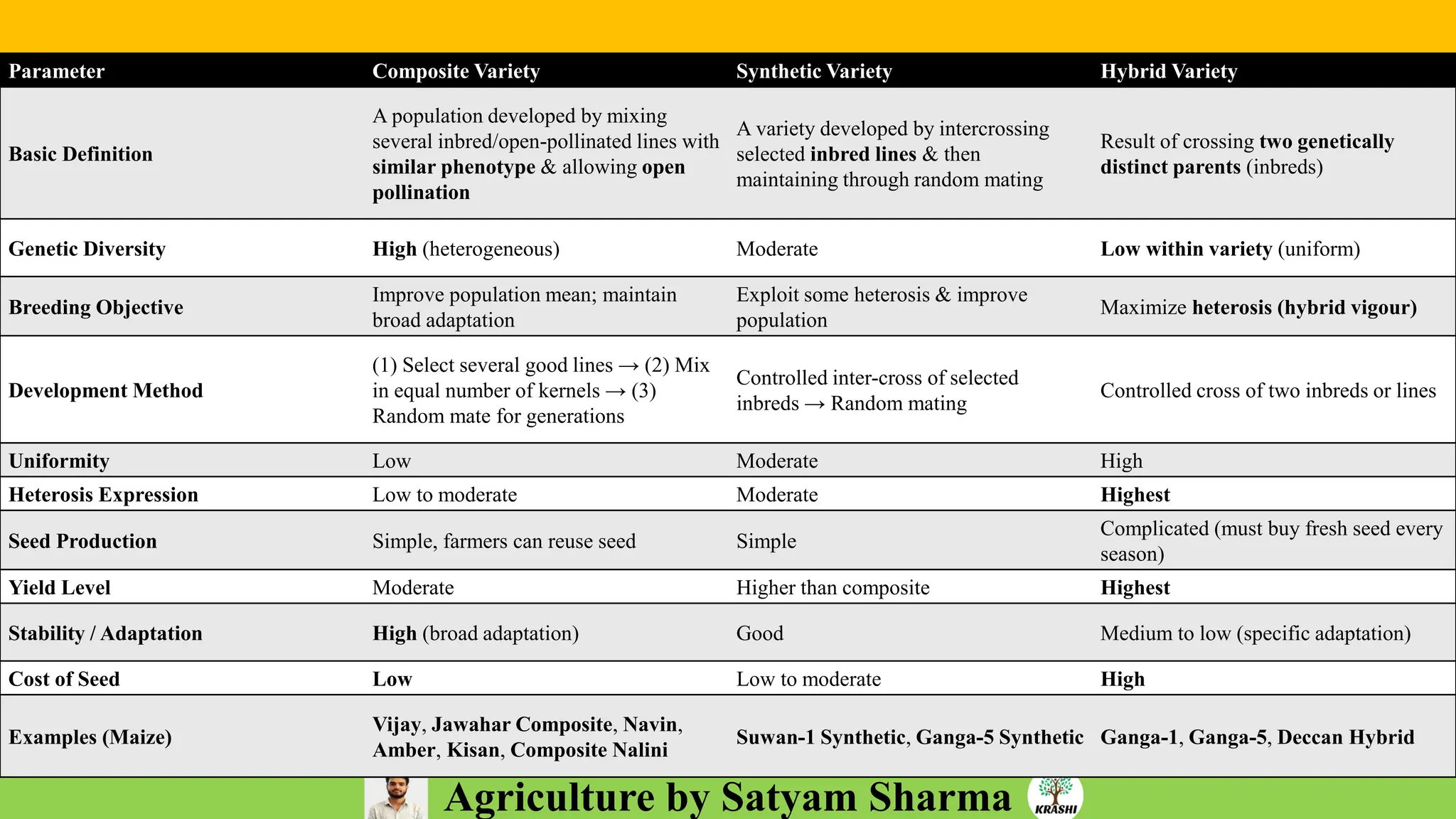
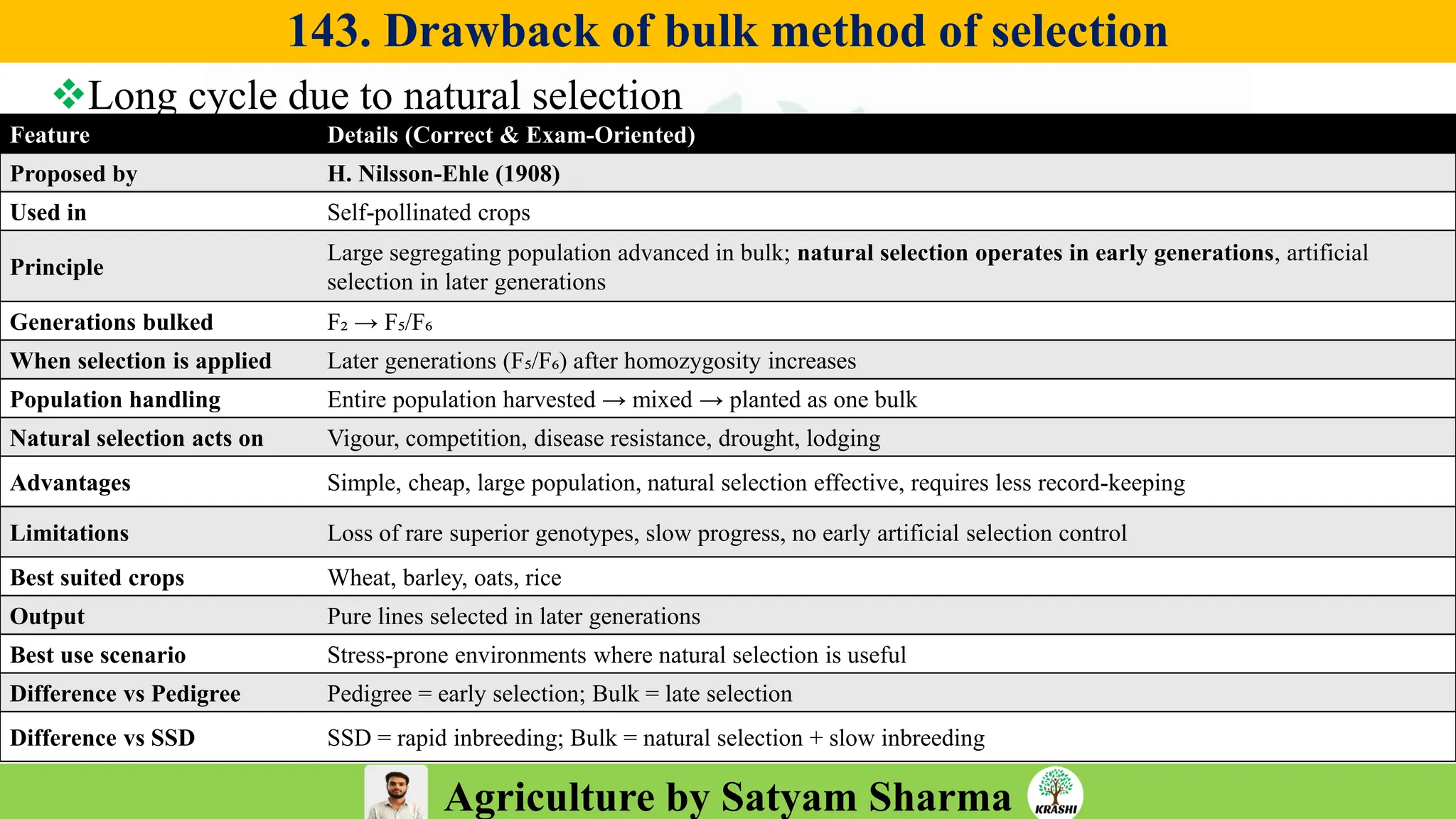
– Plant Breeding (Hybridization, Heterosis, DUS testing)
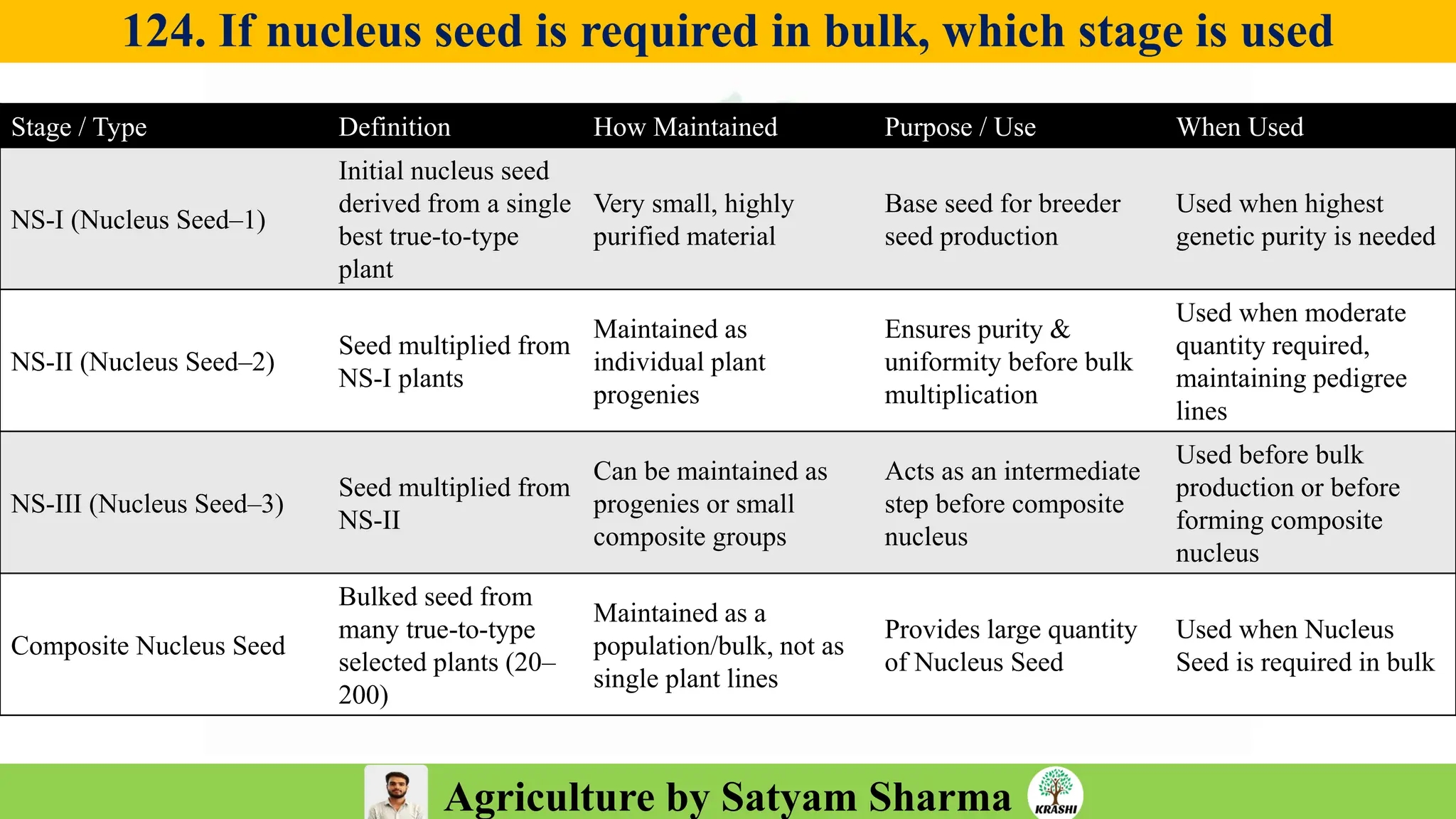
– Seed Science (Vigor, Tetrazolium, Genetic purity)
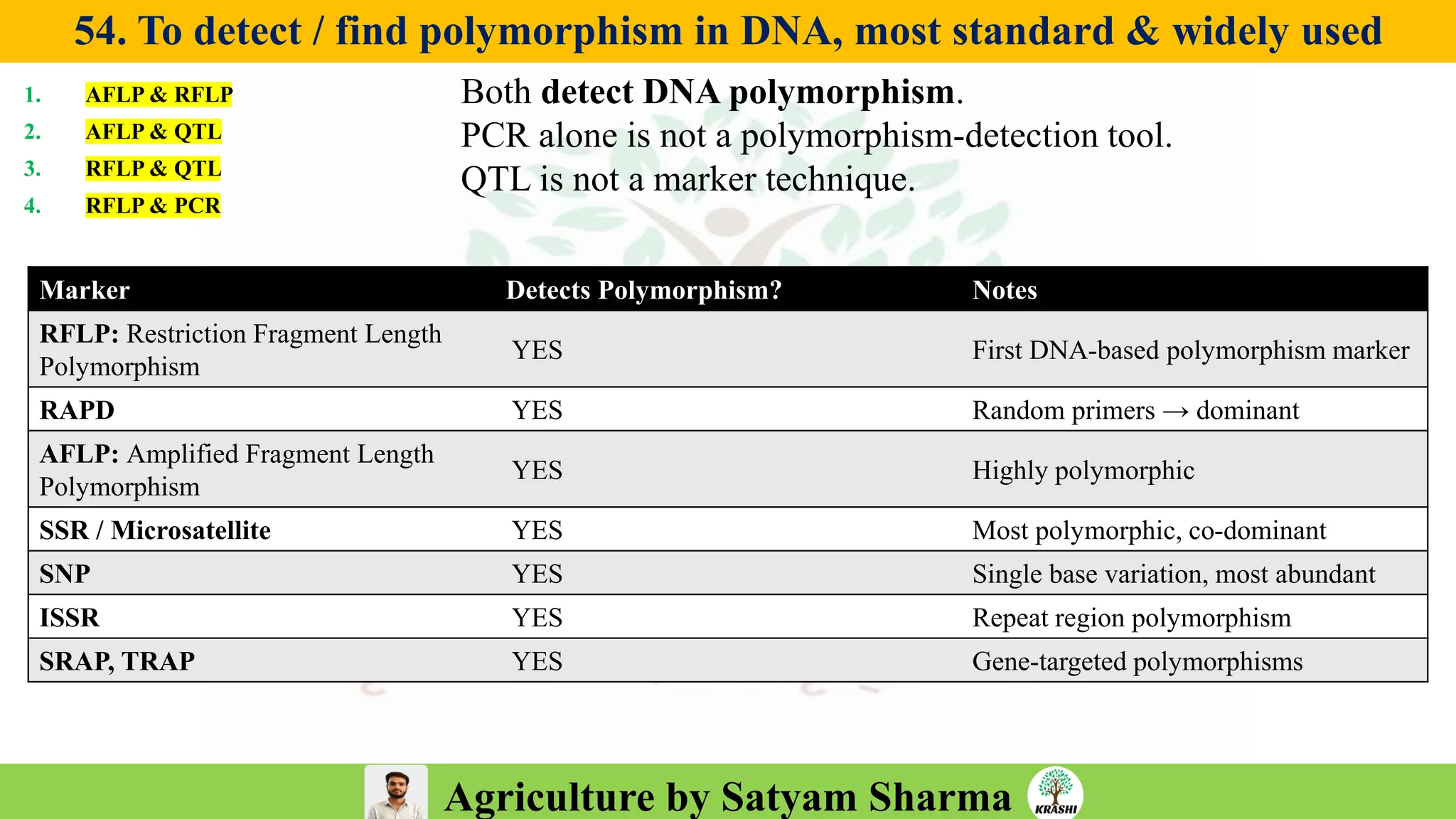
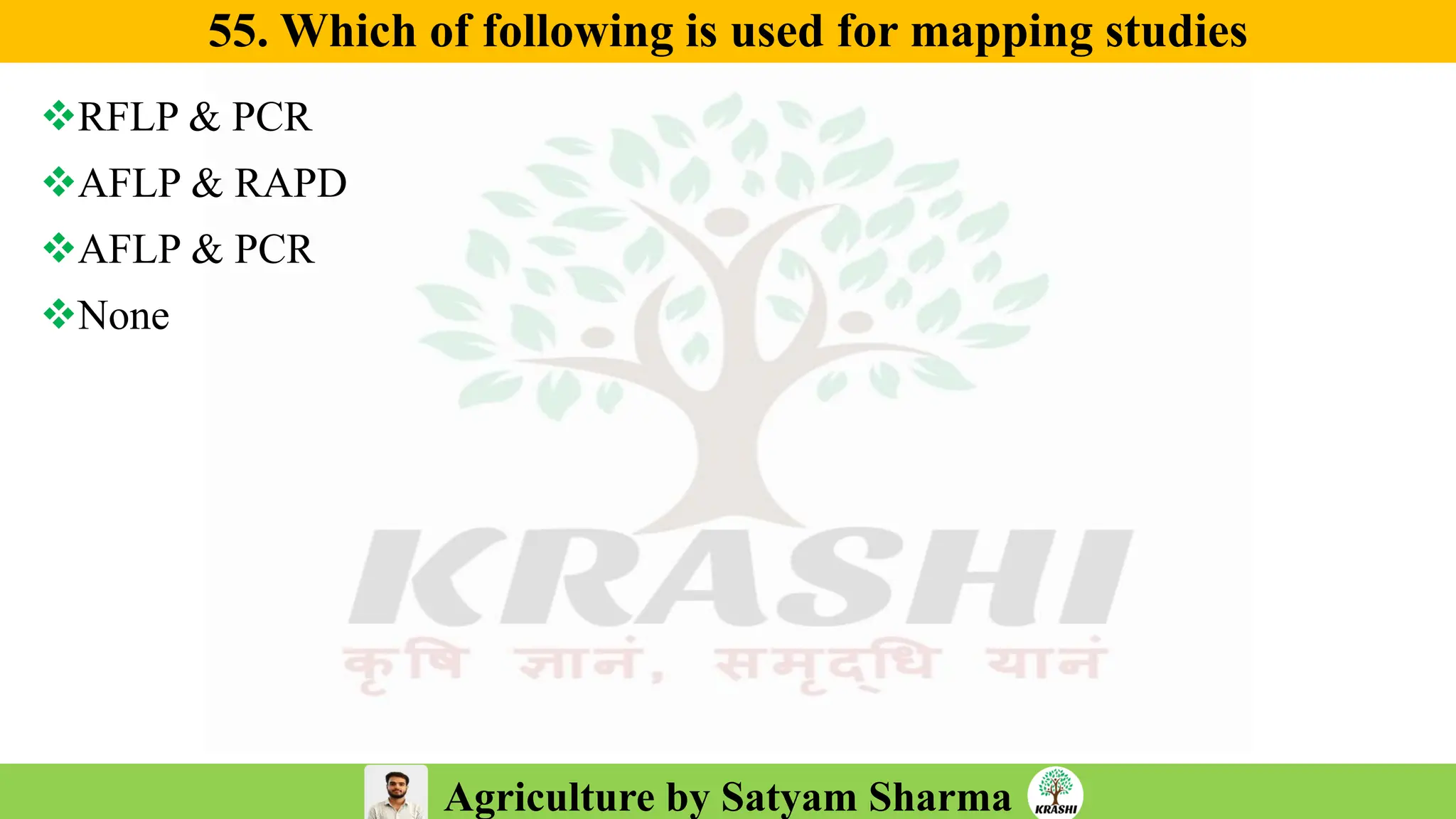
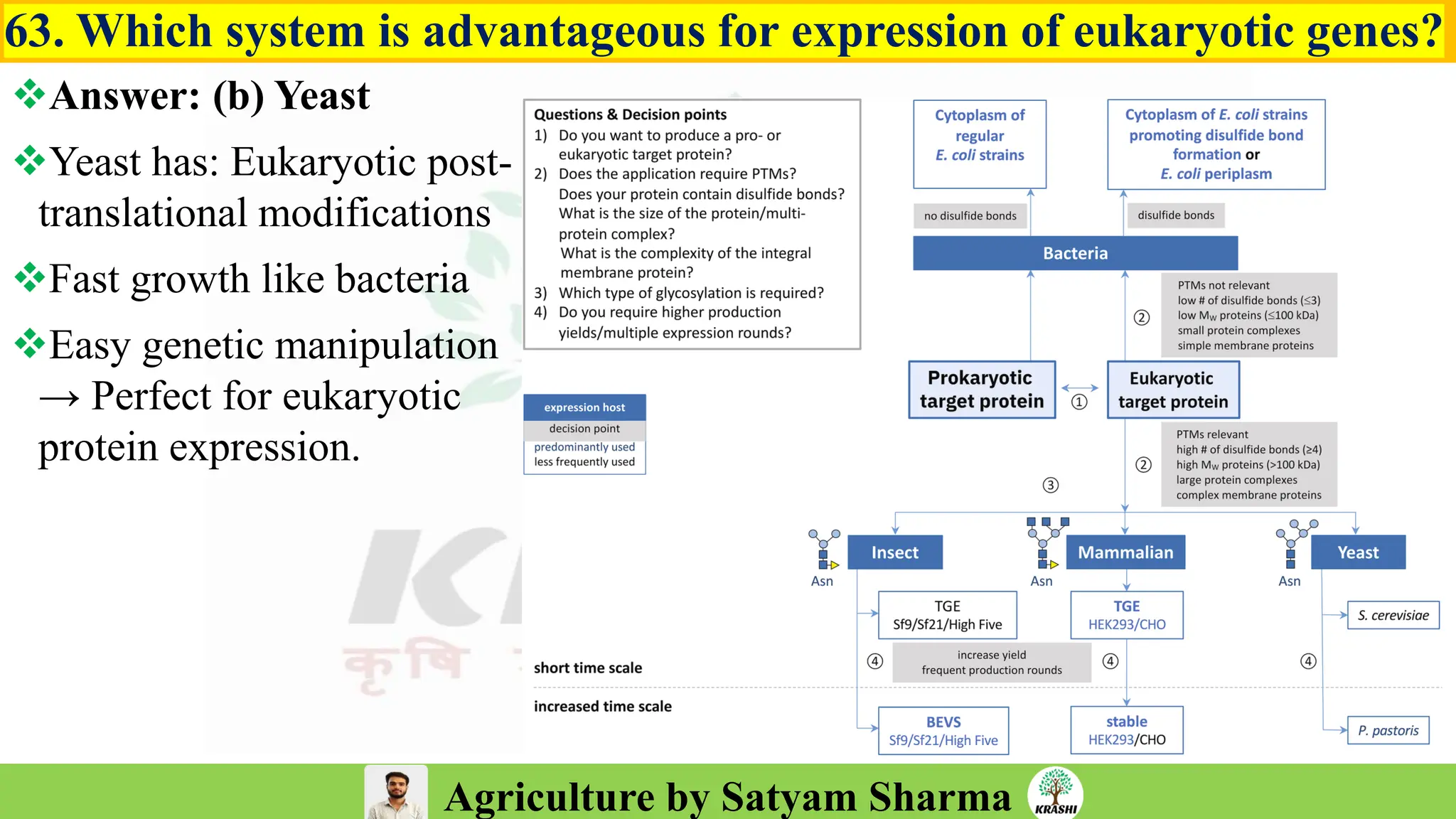
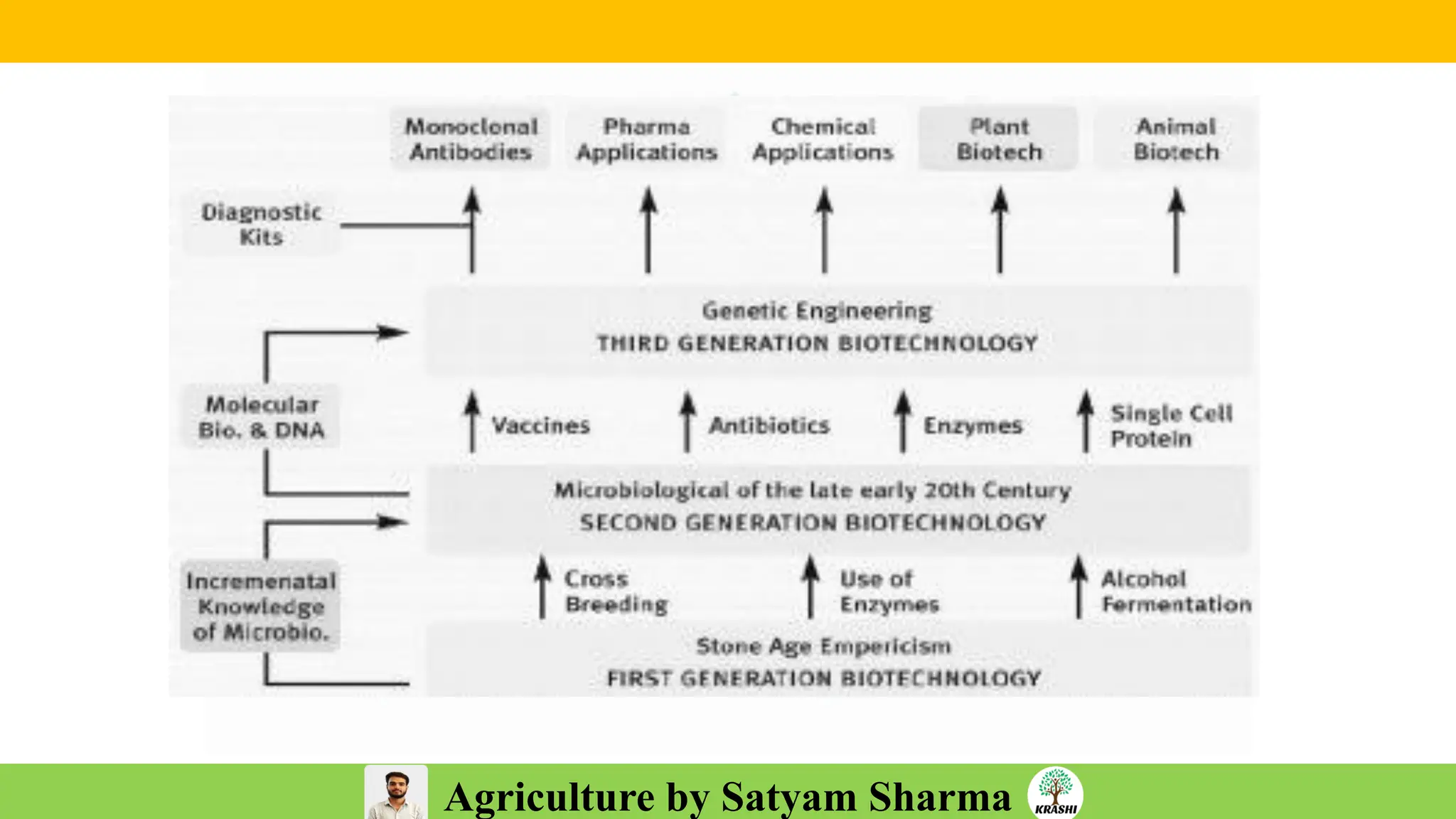
– Biotech (Markers, SSR, SNP, QTL mapping)
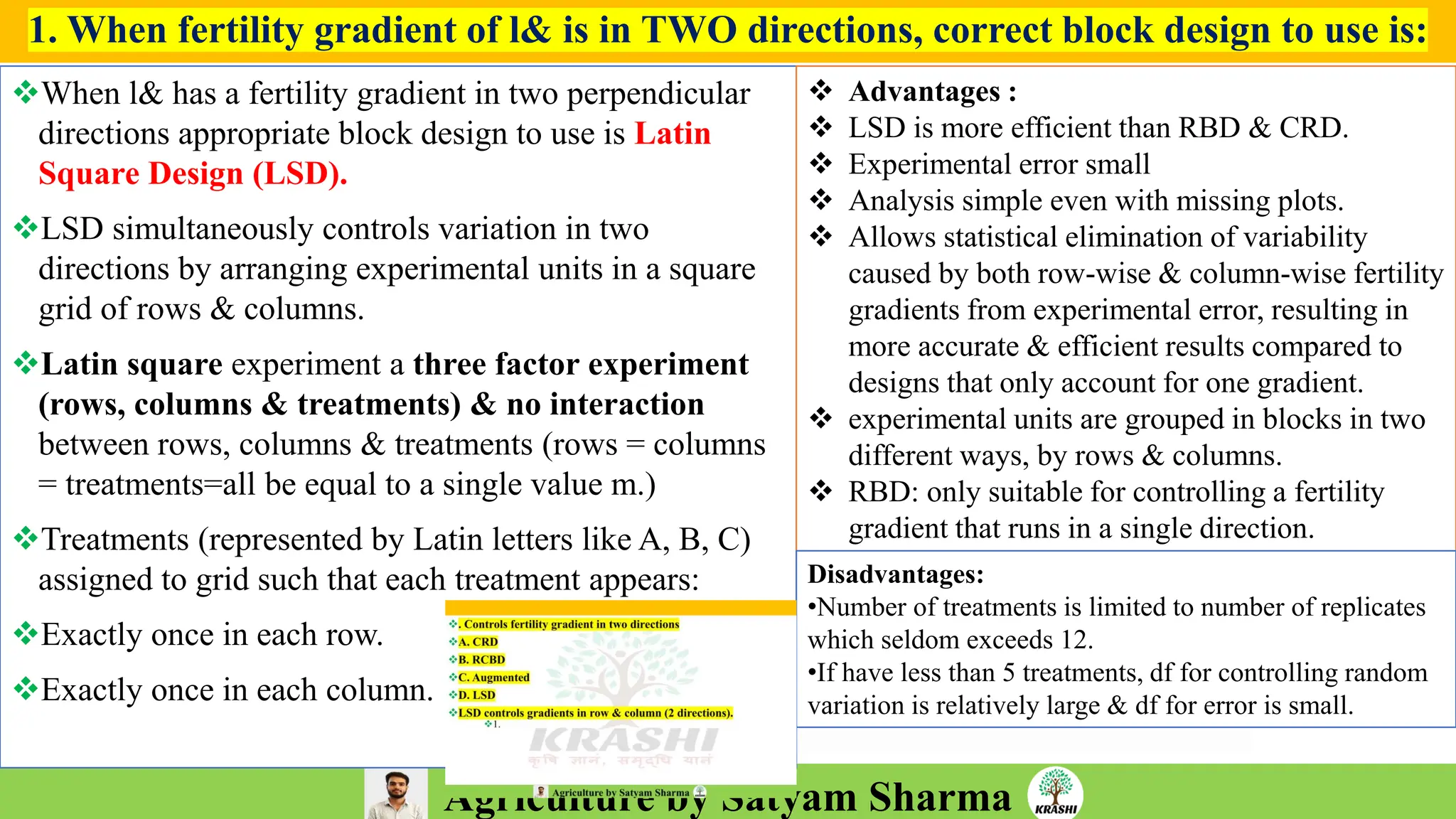
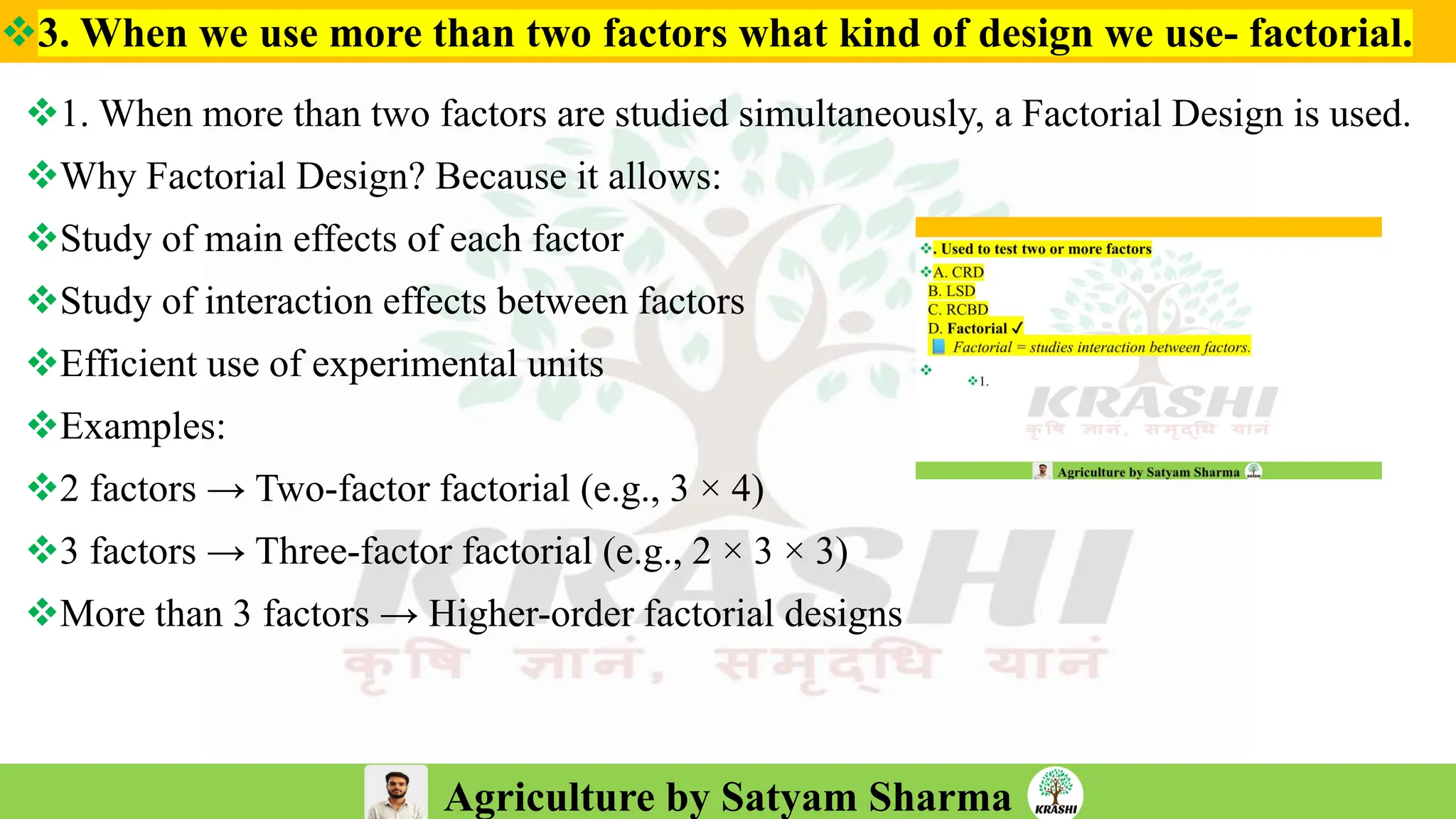
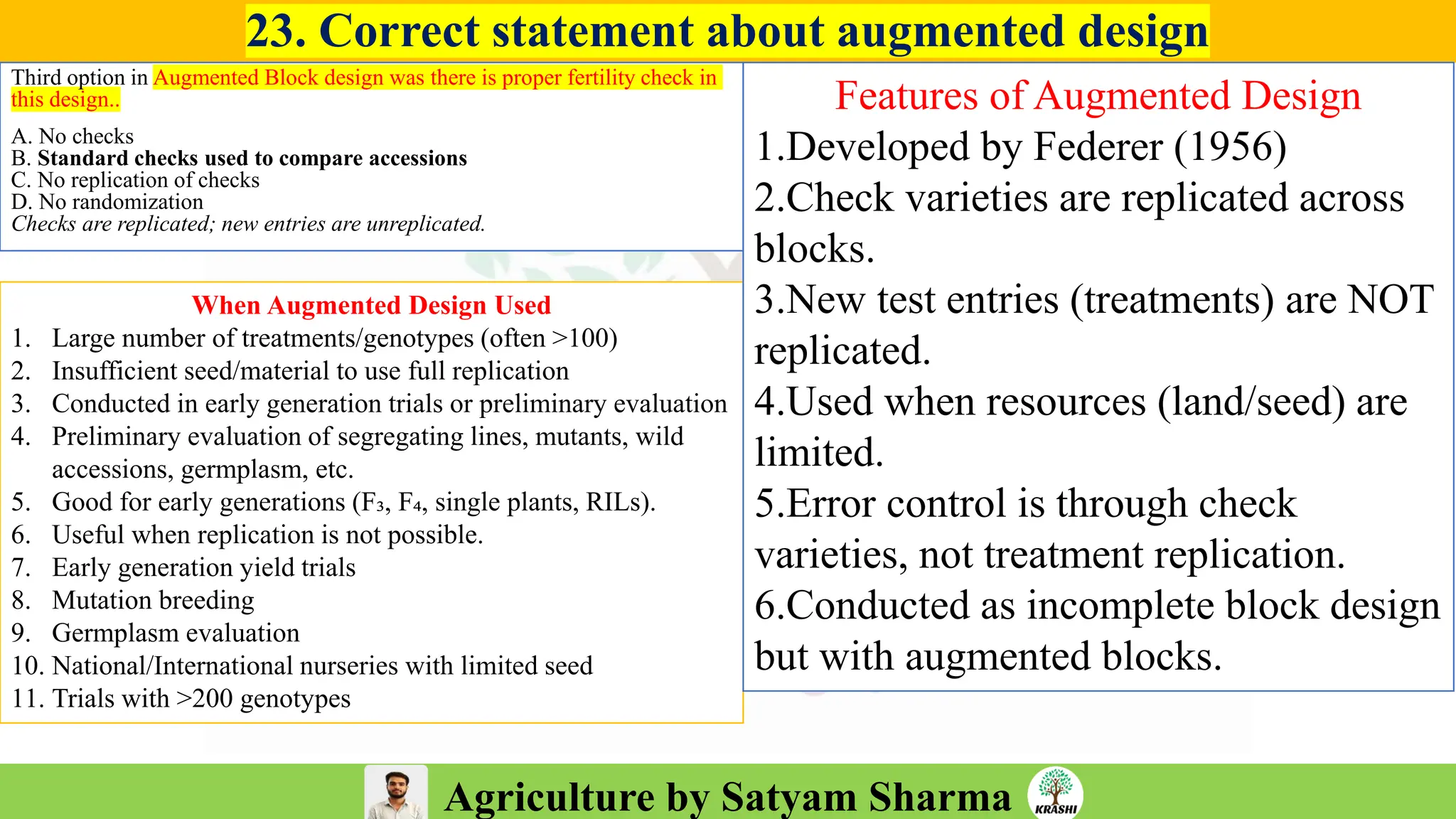
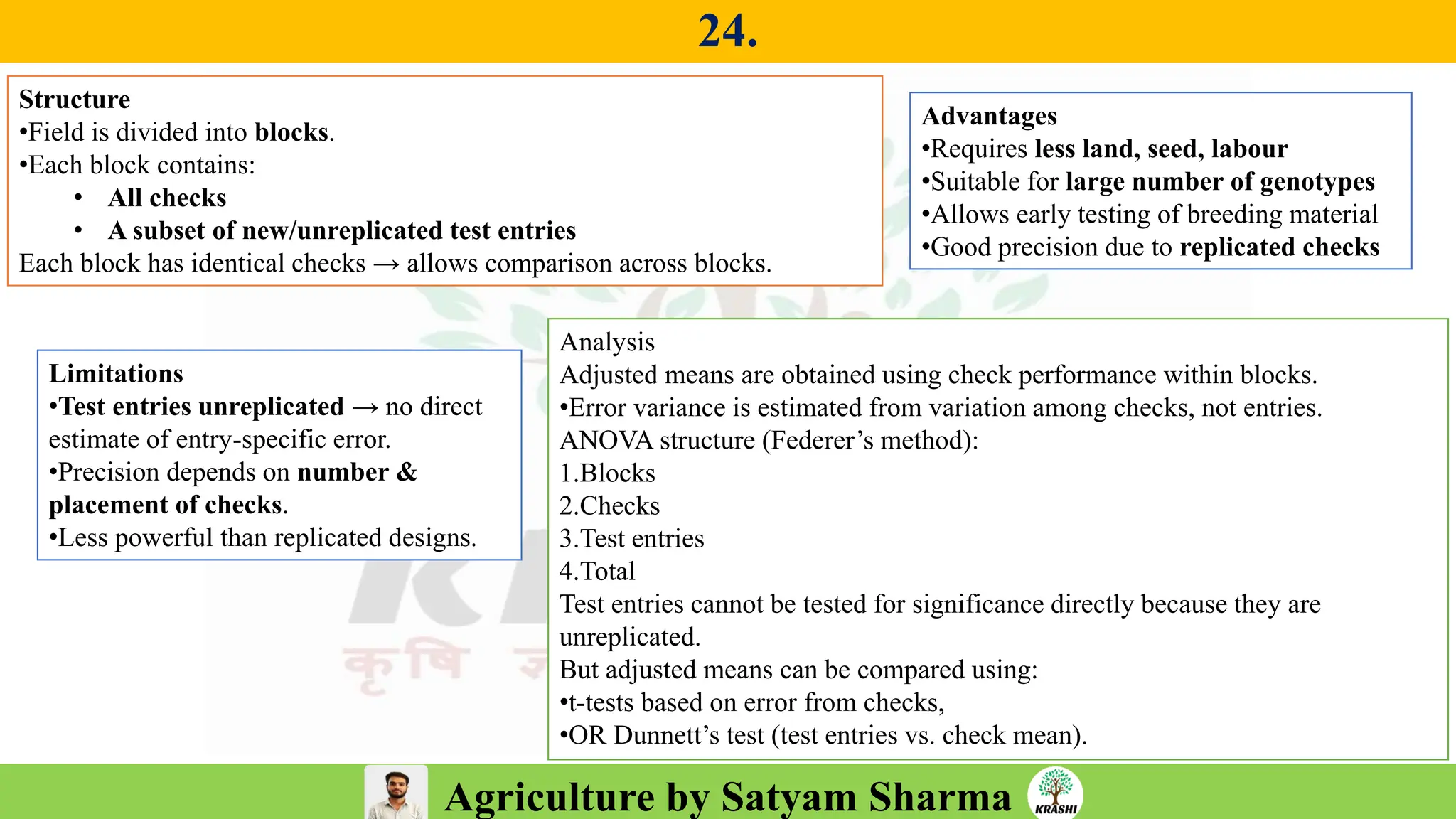
– Experimental Designs (RCBD, LSB, Augmented Design)
• Toughest section
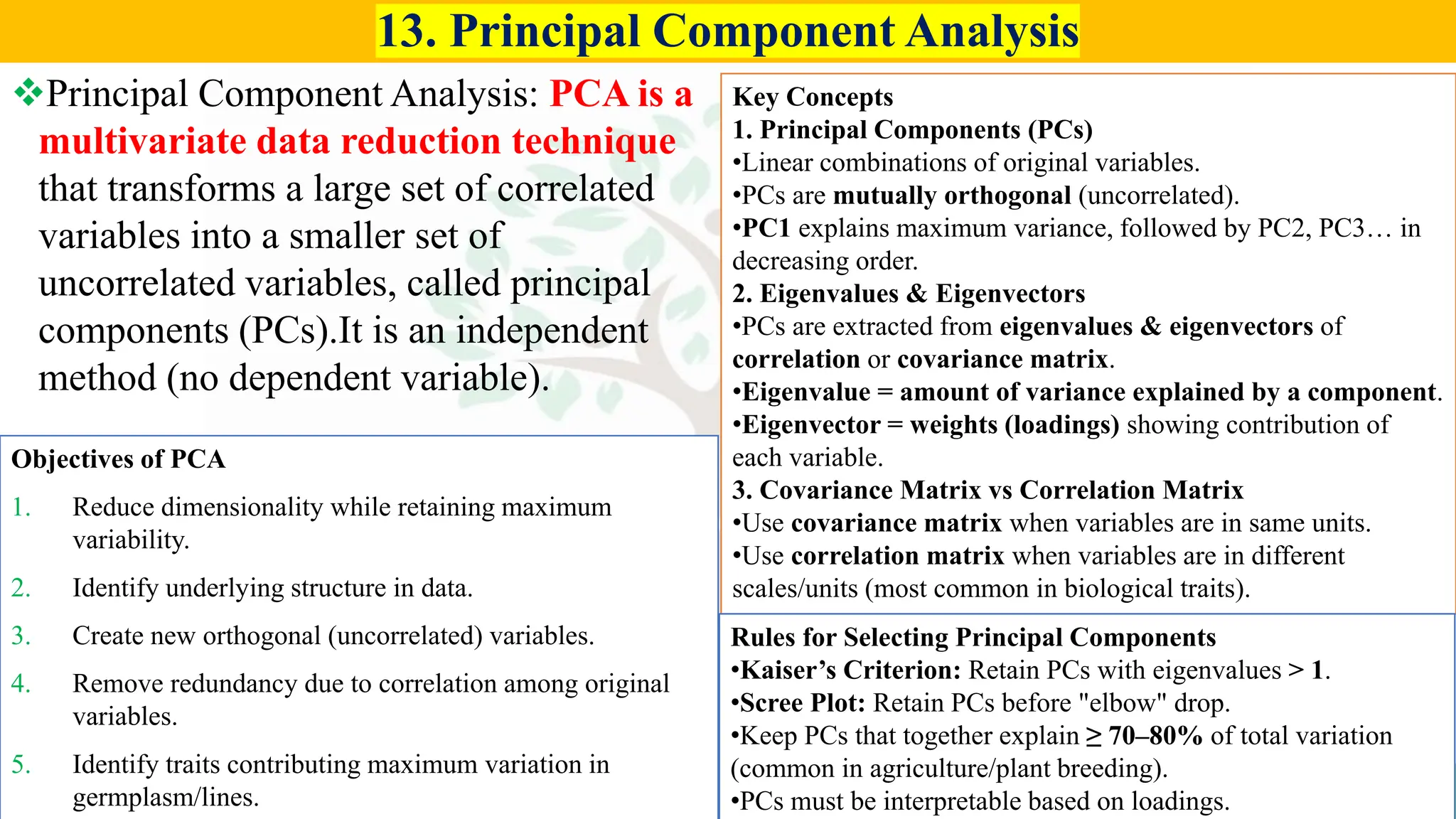
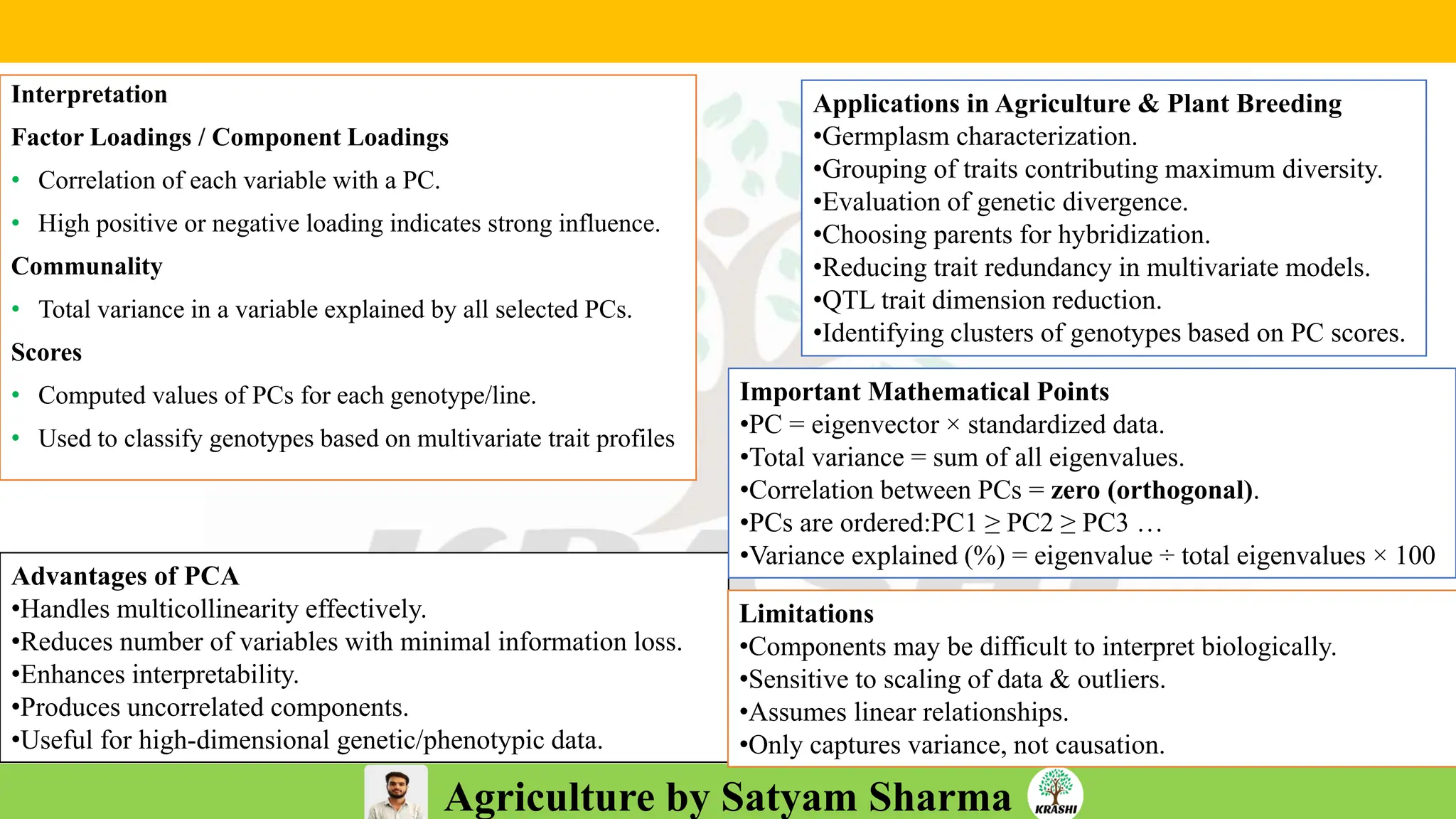
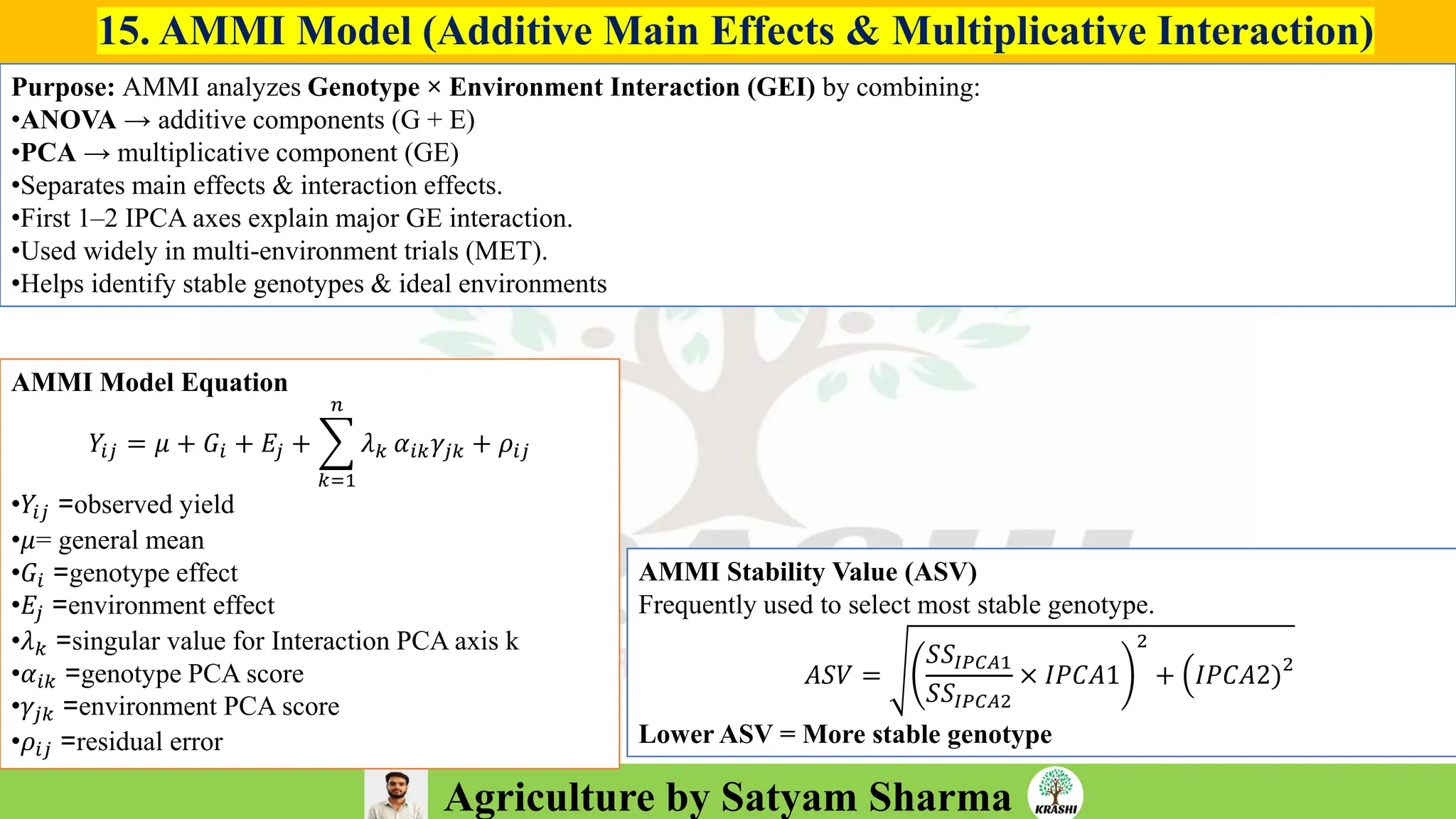
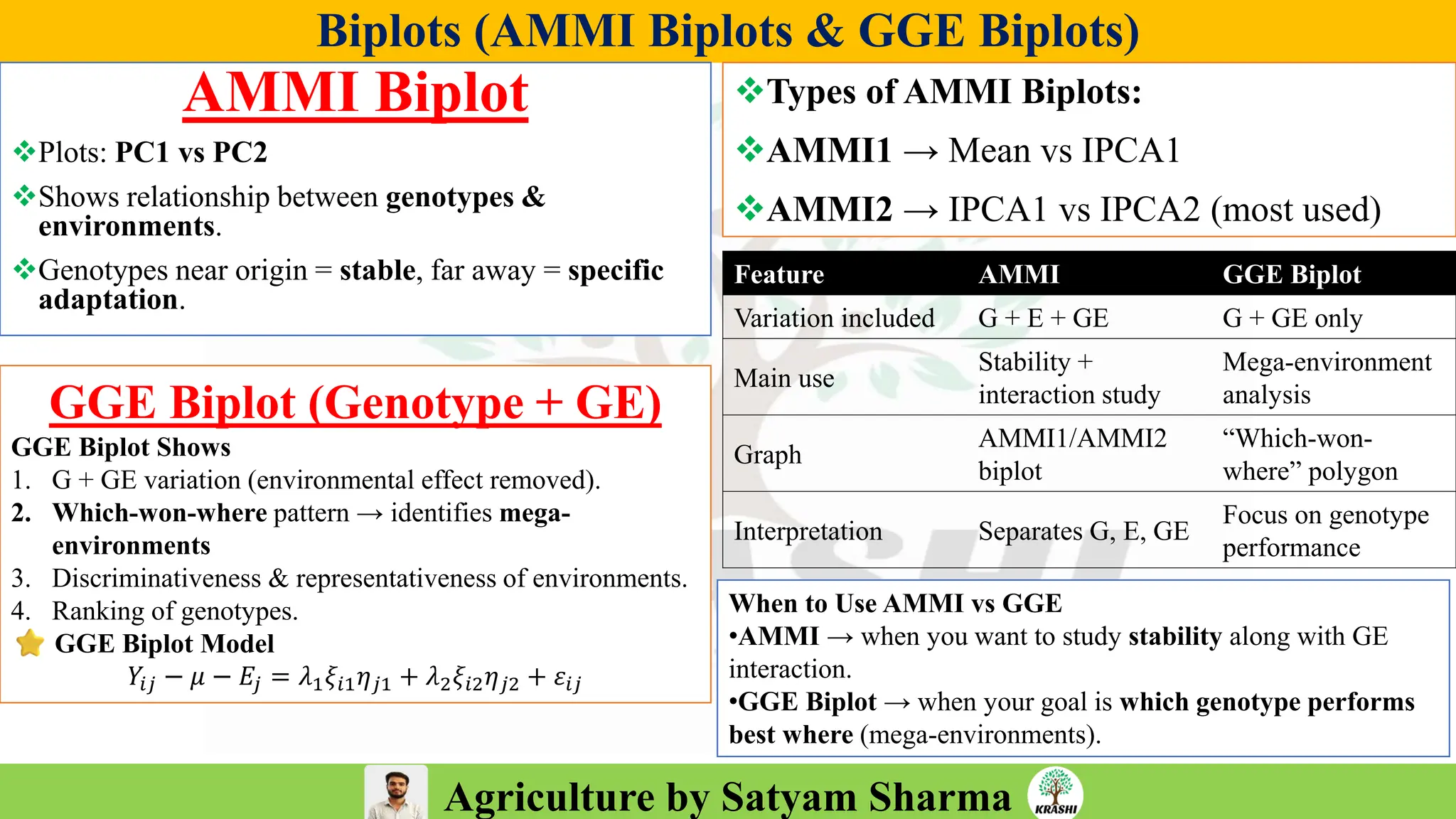
• Conceptual questions from:
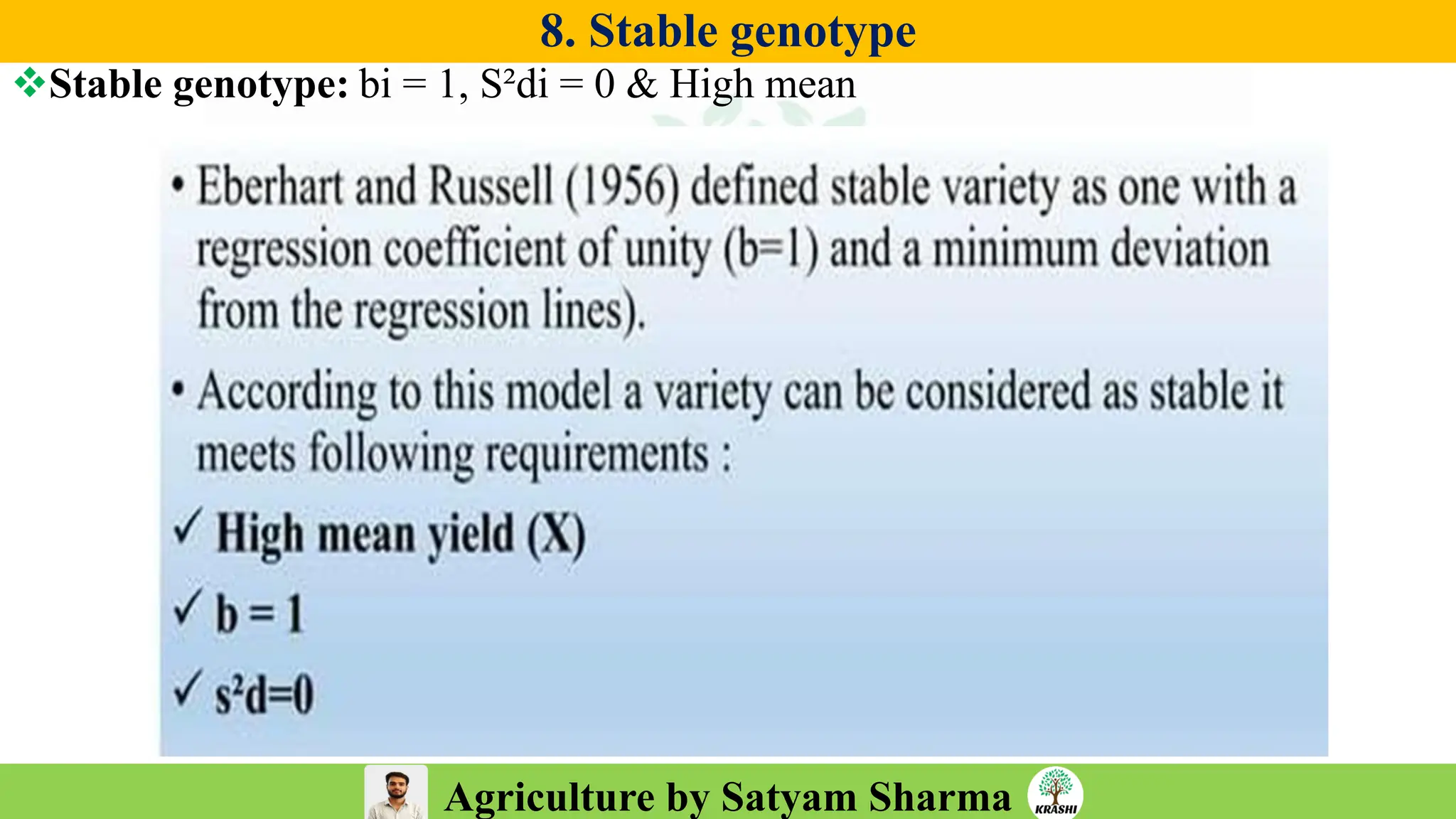
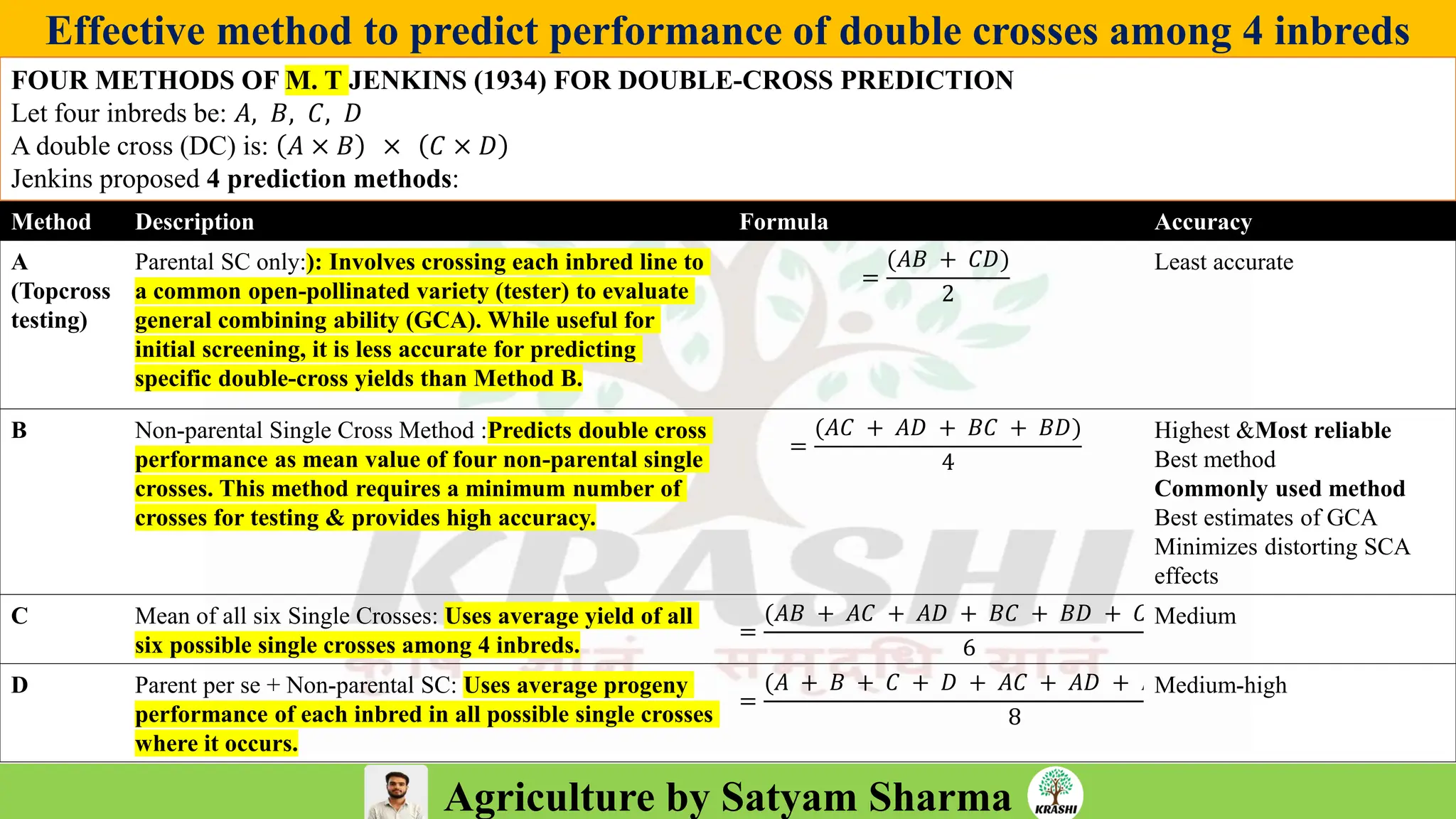
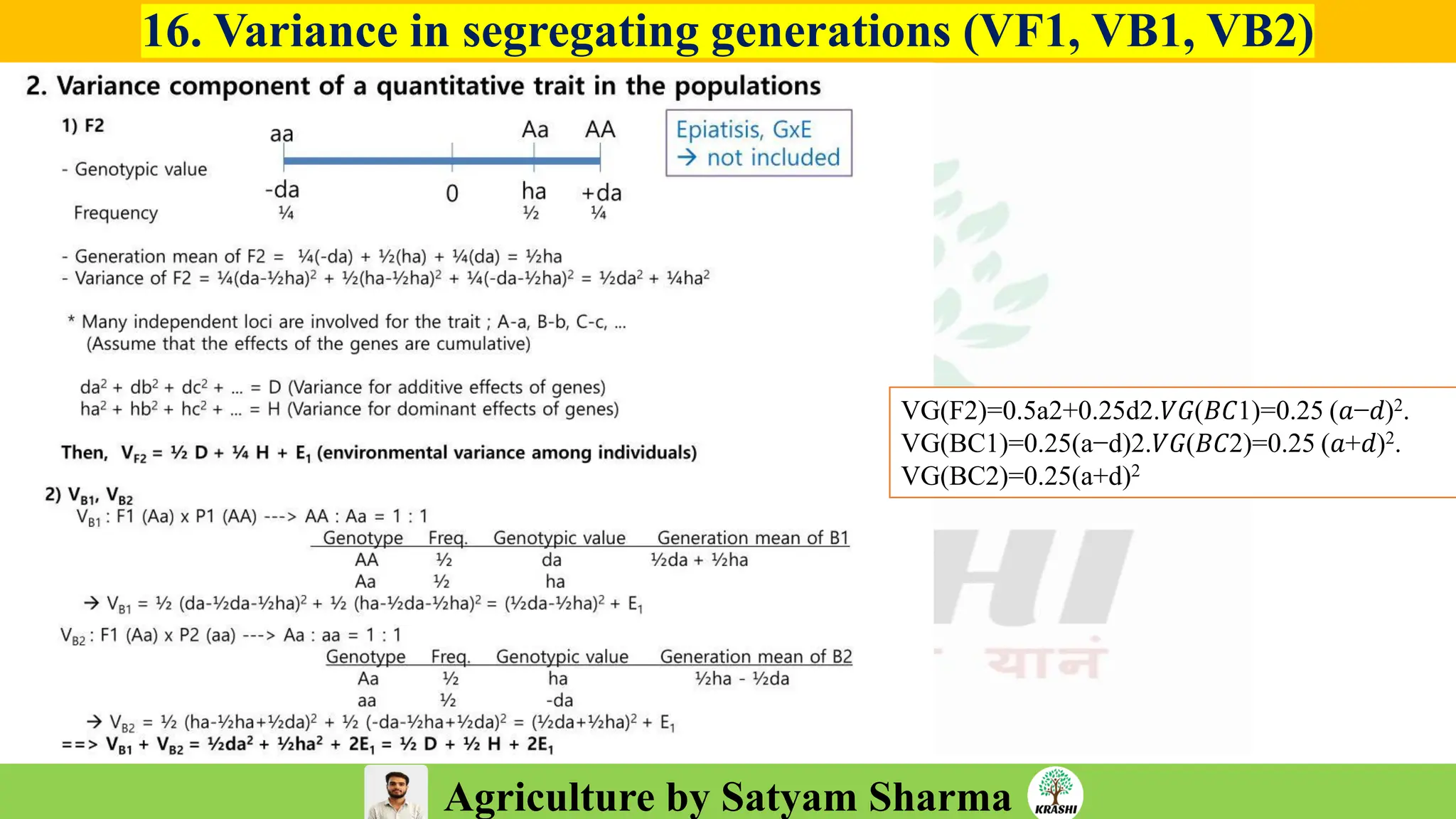
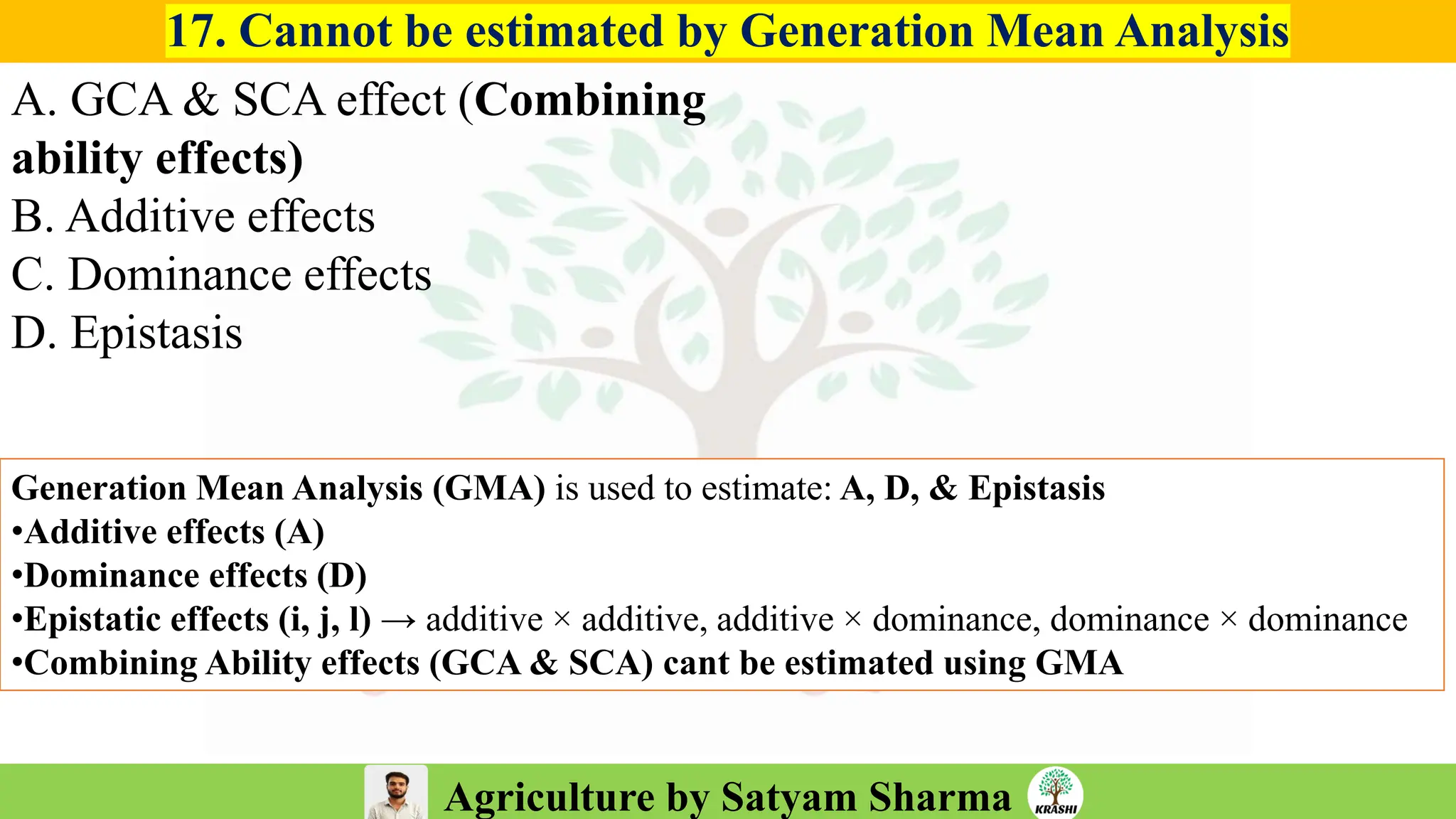
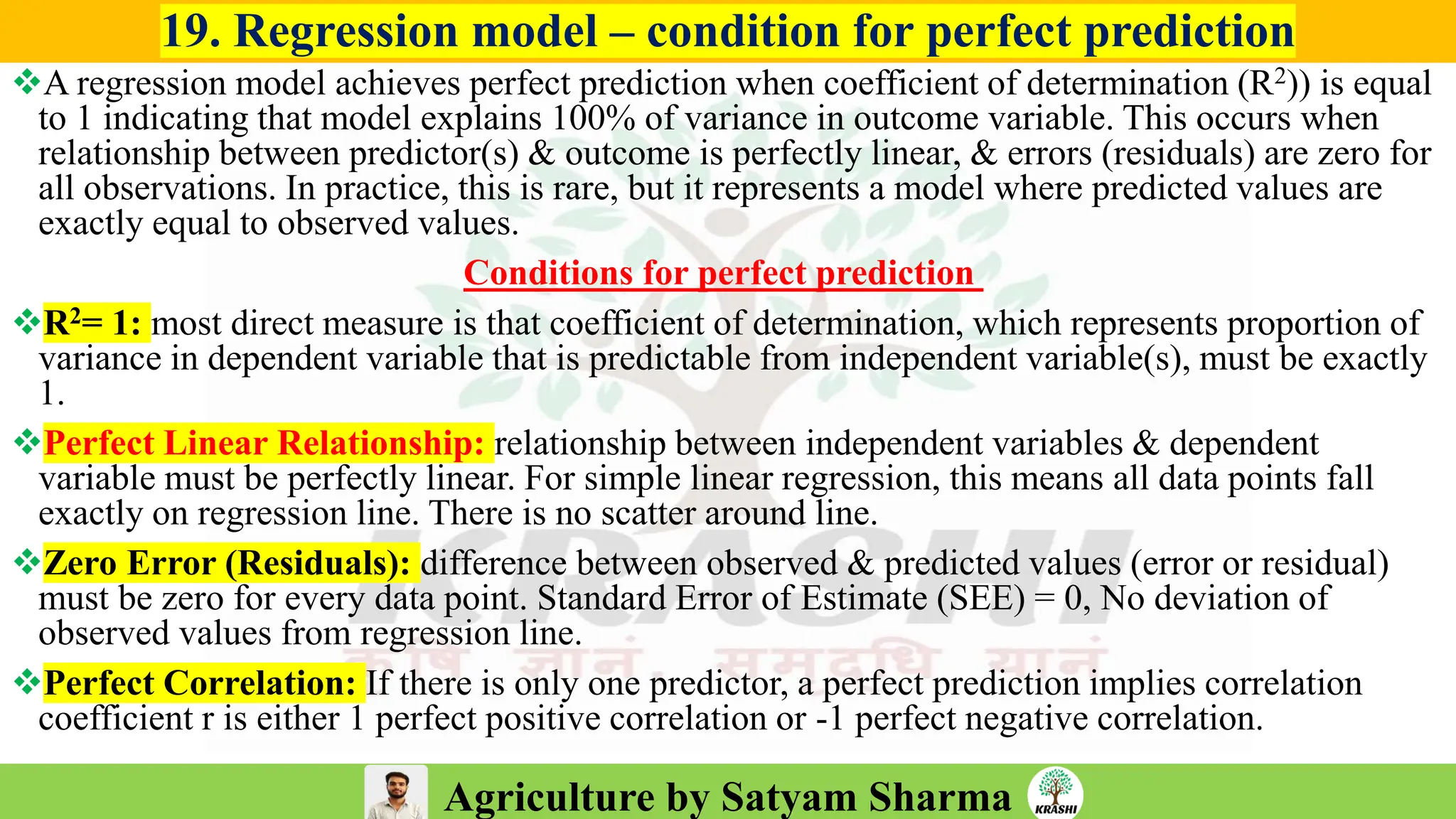
– Generation mean analysis
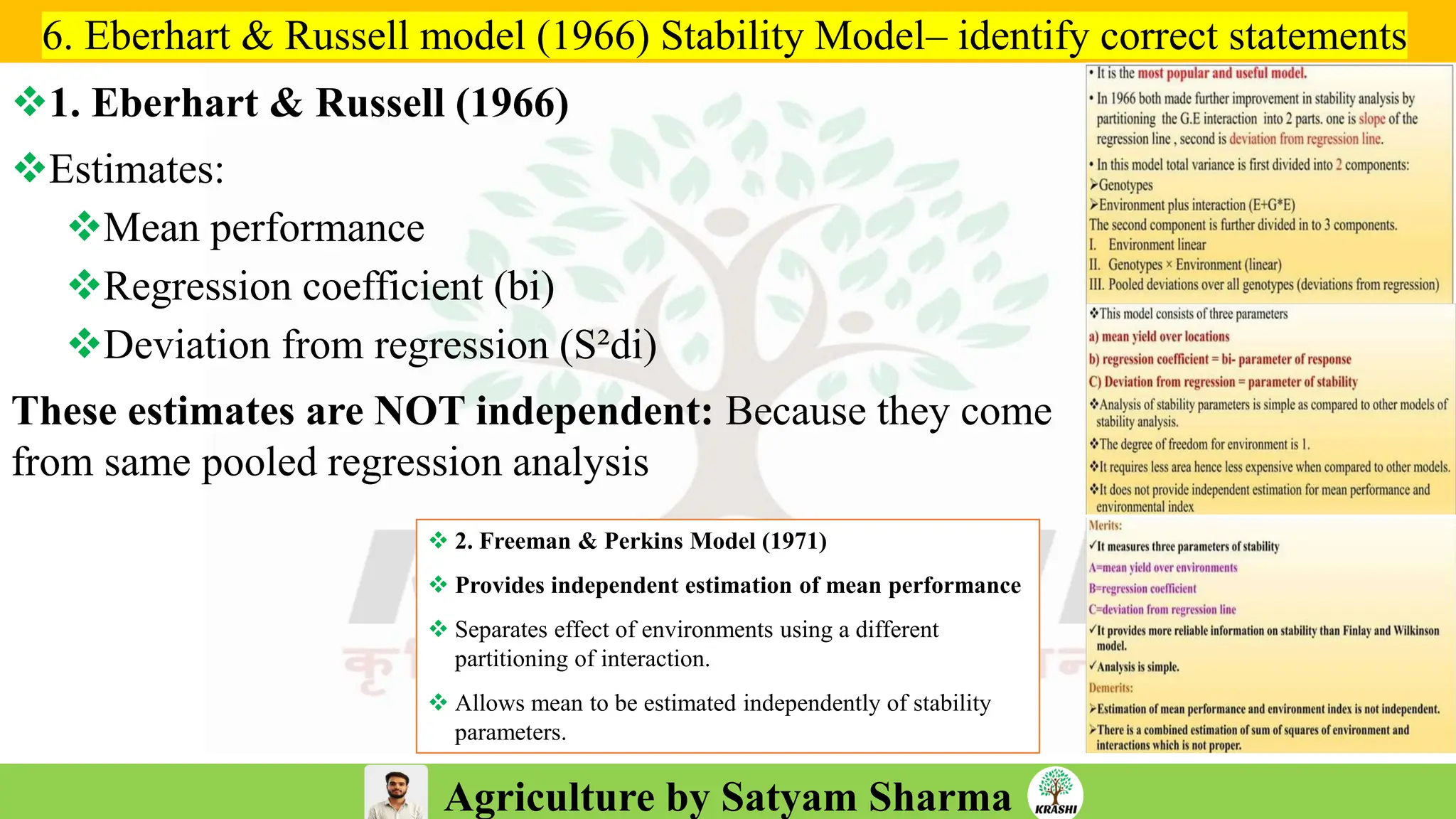
– Stability analysis (Eberhart–Russell, AMMI, GGE)
– Combining ability & diallel
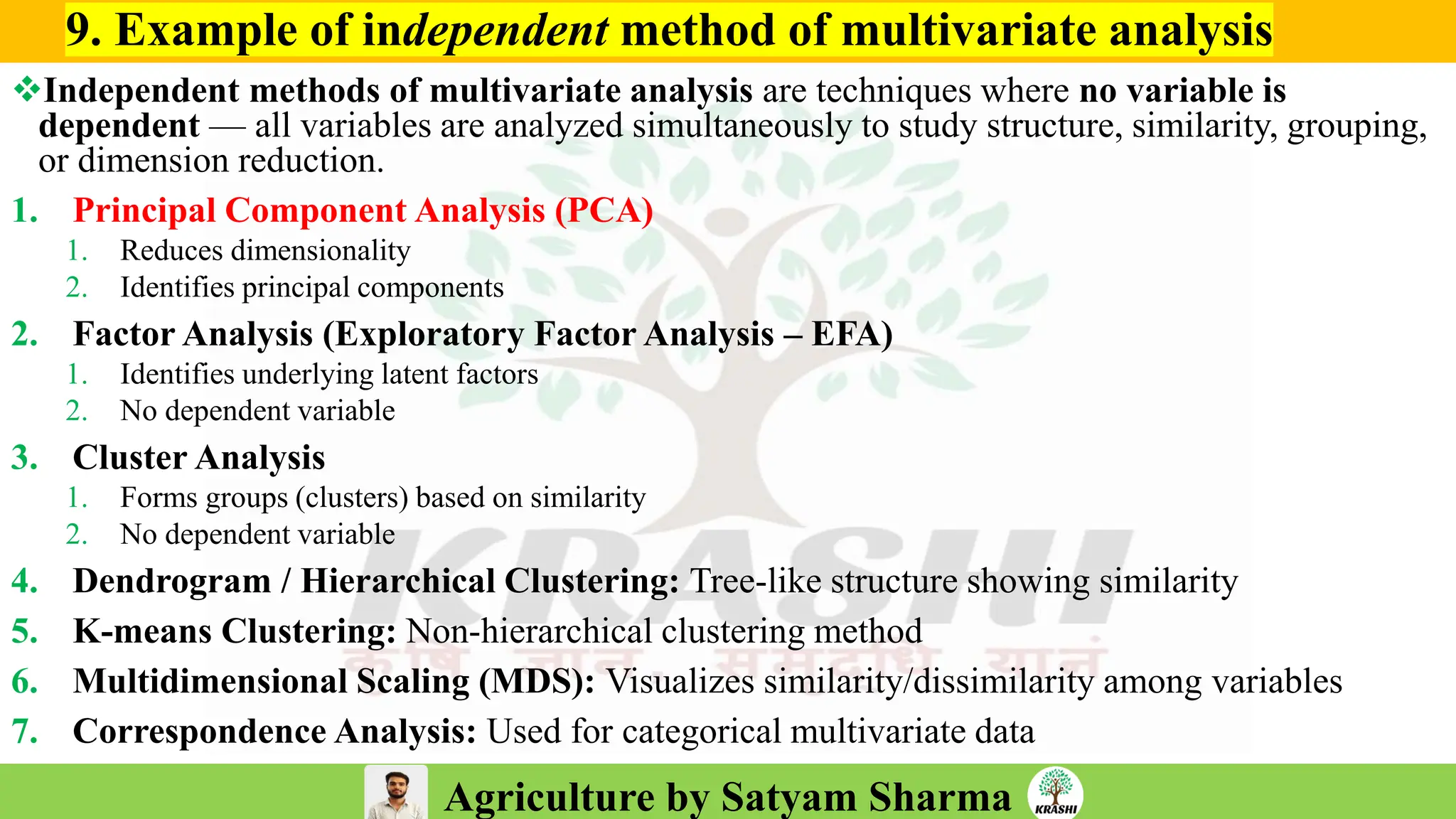
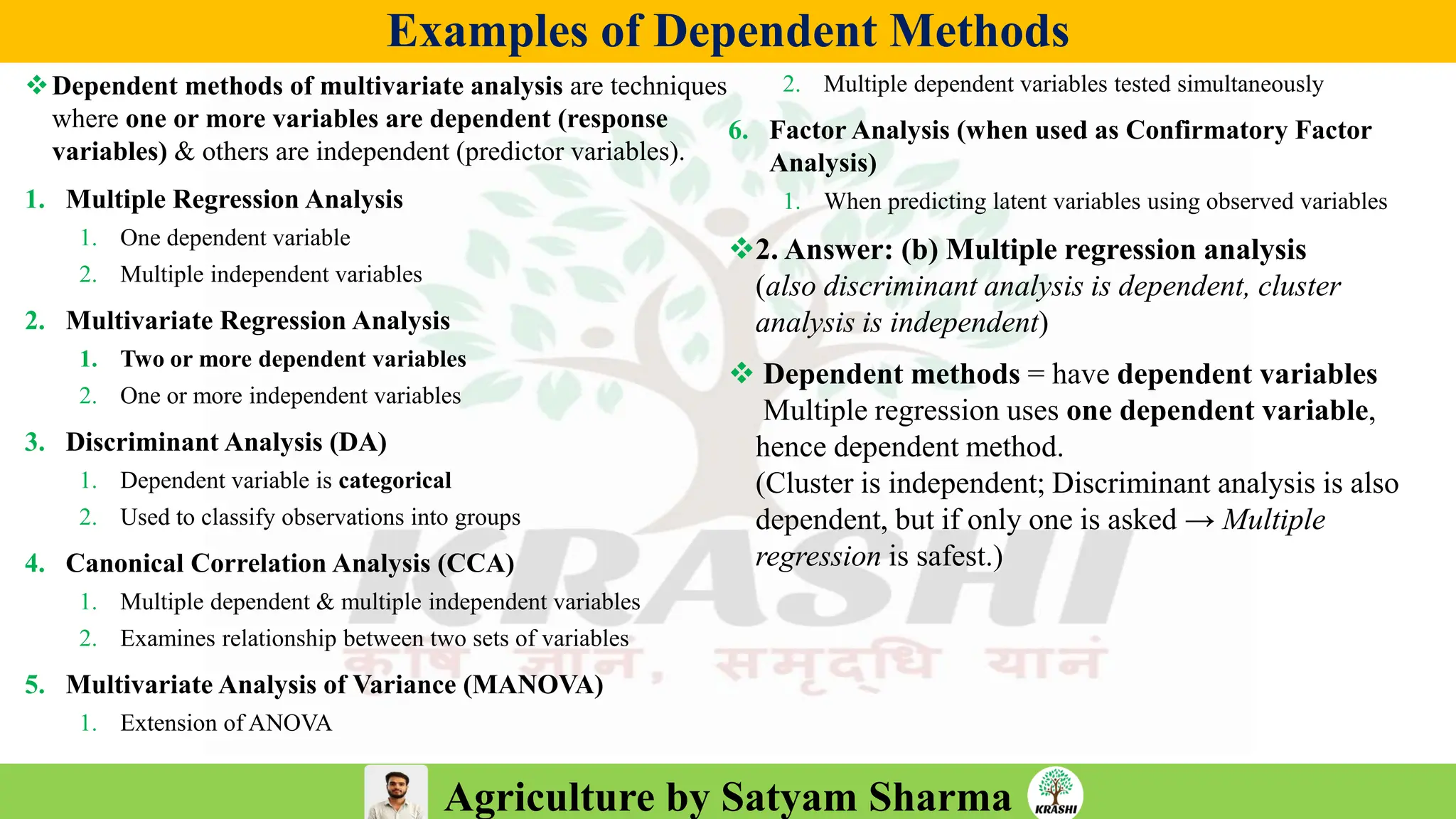
– Molecular breeding
🎯 Who Should Watch This Video?
✔ ASRB NET Aspirants
✔ PhD Entrance candidates
✔ JRF/SRF/ARS preparation students
✔ Agriculture, Genetics, Plant Breeding, Seed Science students
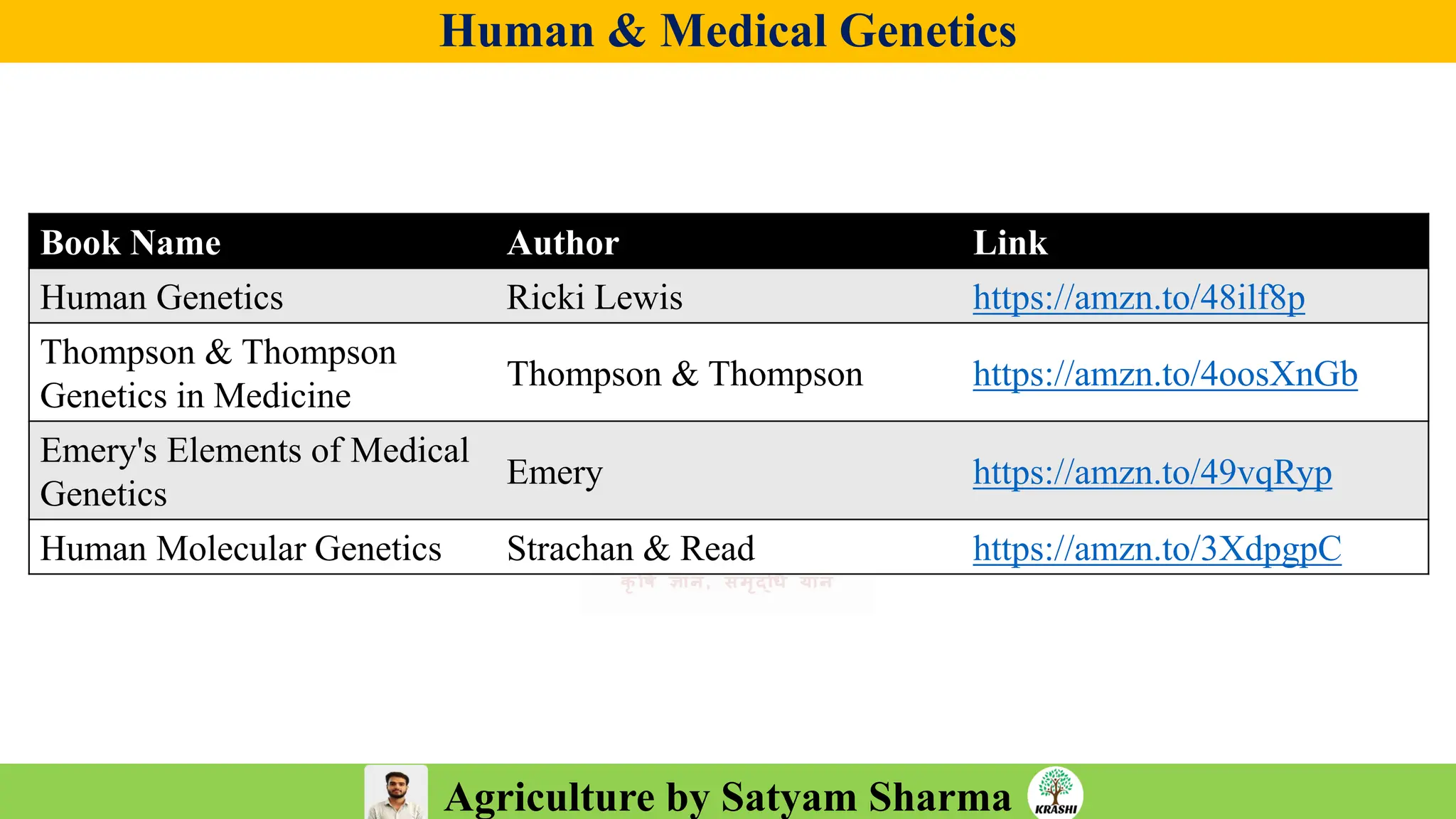
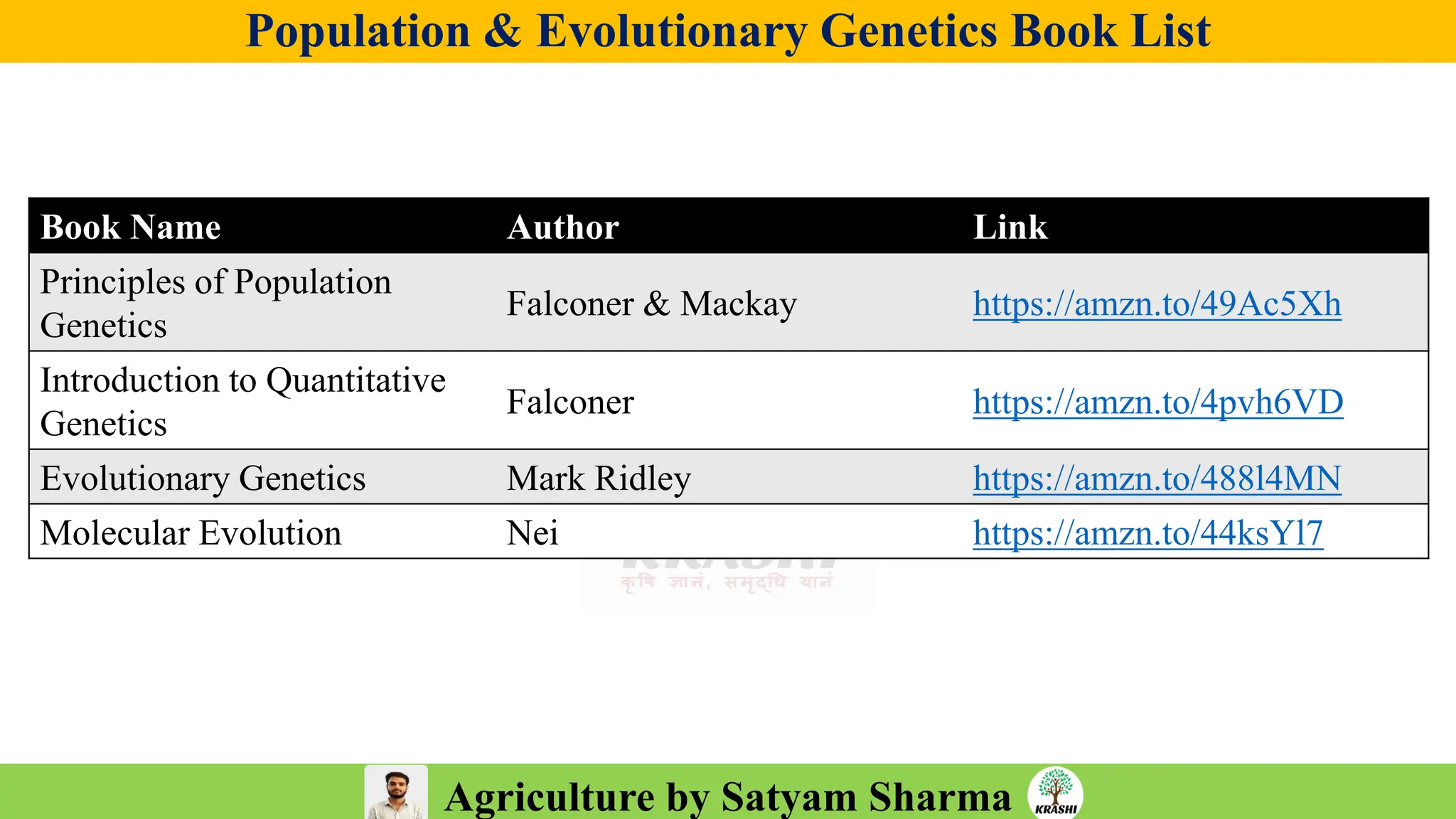
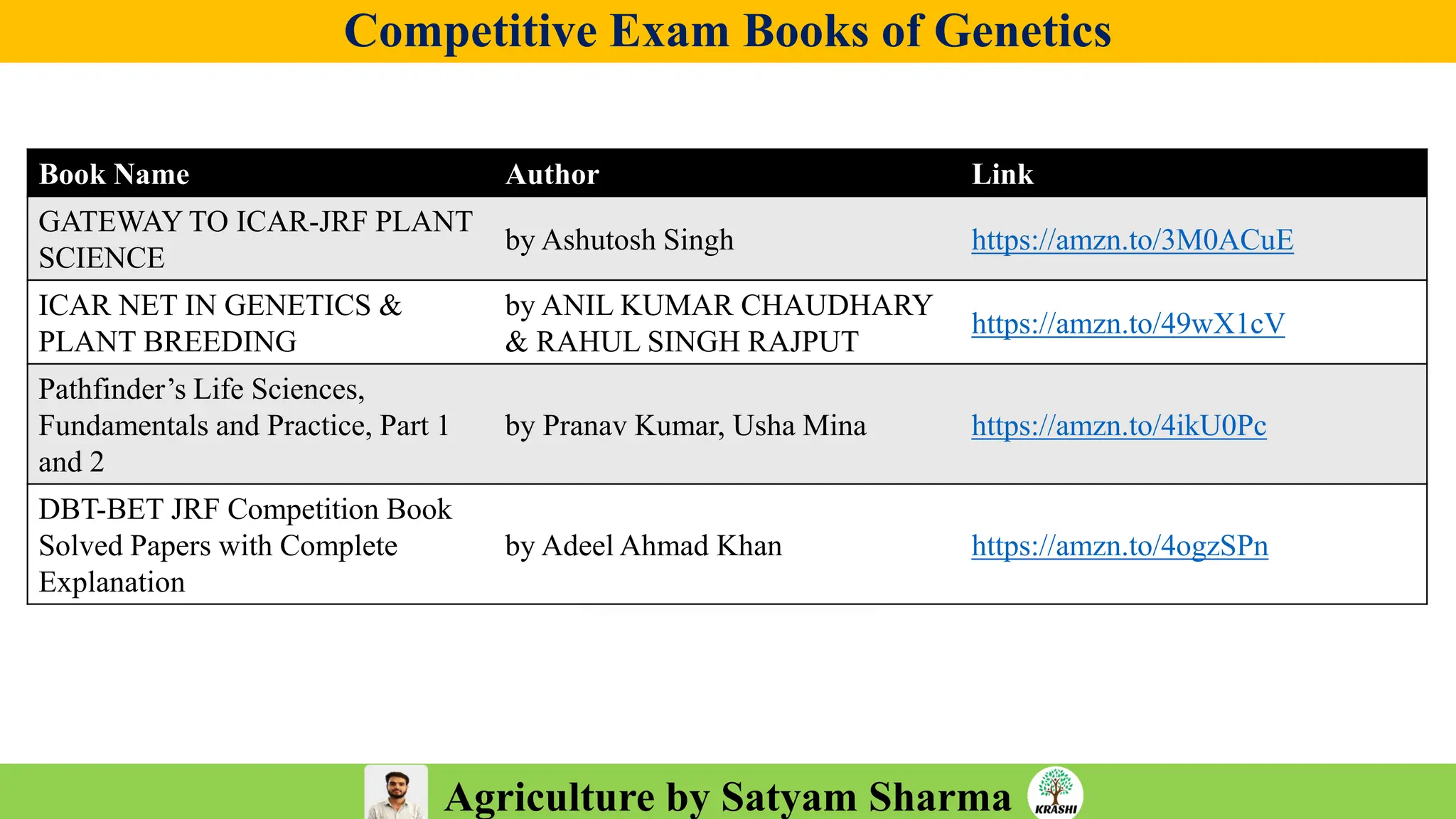
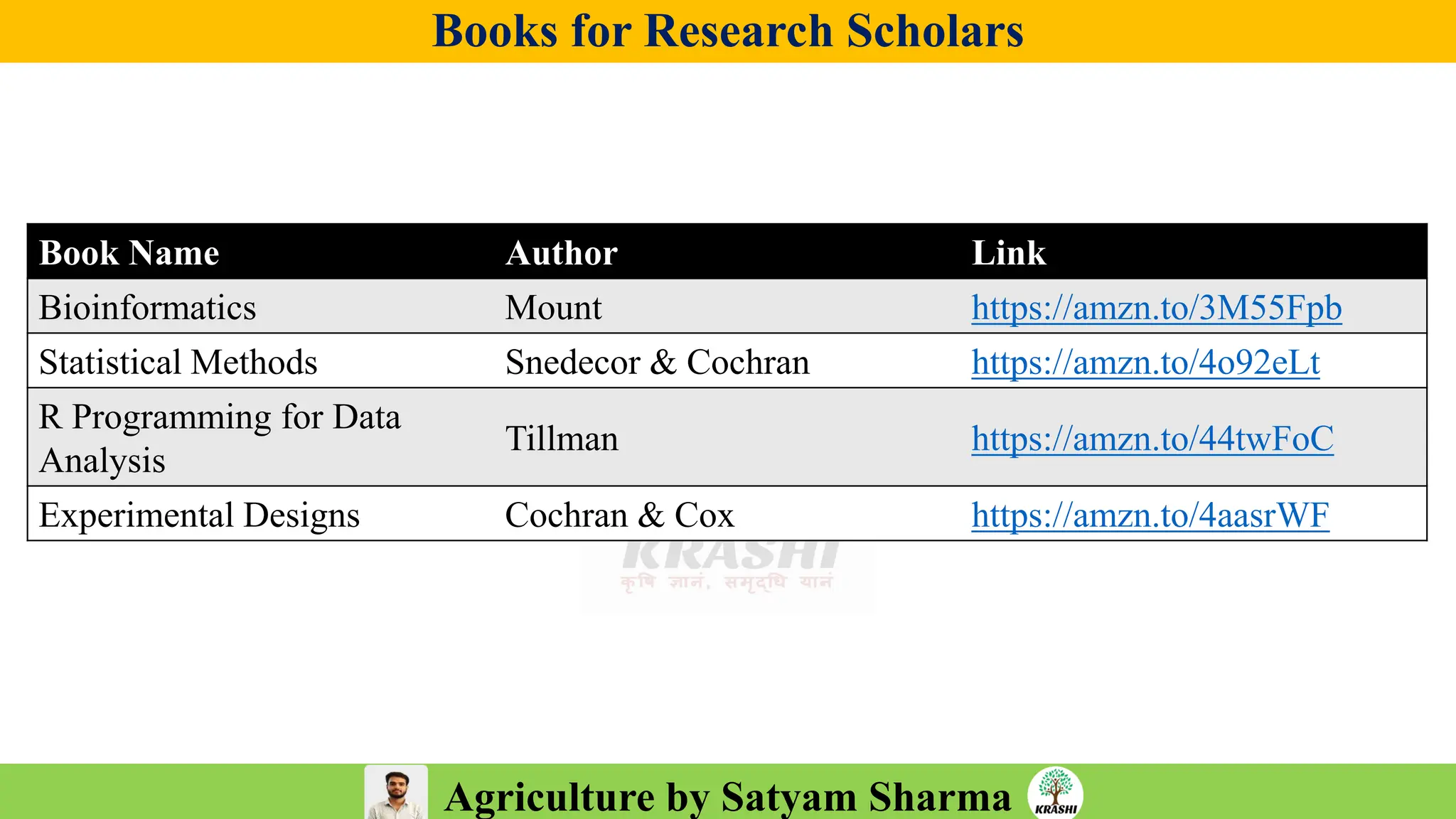
• PDF with important topics
• Topic-wise MCQs
• Previous year trends
• 2025 full recall question
✔ Complete ASRB NET 2025–26 coaching
✔ Free mock tests
✔ Chapter-wise MCQ series
✔ Live doubts & revision sessions
ASRB NET 2025 paper discussion
ASRB NET 2025 analysis
ASRB NET answer key 2025
ASRB NET agriculture 2025
ASRB NET solution 2025
ASRB NET plant breeding 2025
ASRB NET seed science 2025
ASRB NET 2025 cutoff prediction
ASRB NET 2025 question paper
ASRB NET preparation 2025
ASRB NET agriculture exam analysis
ASRB NET plant breeding exam 2025
#JRF2026 #PlantScience #SatyamSharma #ICARAIEEA #PlantBreeding #Genetics #SeedScience #Biotechnology #AIEEAPG #JRFPreparation #SatyamSir
#PlantBreeding #Domestication #GermplasmConservation #SatyamSharma #AgricultureExams